Difference between revisions of "Part:BBa K782063"
| Line 2: | Line 2: | ||
<partinfo>BBa_K782063 short</partinfo> | <partinfo>BBa_K782063 short</partinfo> | ||
| − | Additional characterization of BBaI712009 by [http://2016.igem.org/Team:LMU-TUM_Munich Munich 2016]: | + | =Additional characterization of BBaI712009 by [http://2016.igem.org/Team:LMU-TUM_Munich Munich 2016]:= |
| + | |||
| + | [[File:Muc16_SequencingofGMK_TK90BioBricks_001.png|thumb|center|920px| Sequencing results of BioBricks [https://parts.igem.org/wiki/index.php?title=Part:BBa_K2170150 BBa_K2170150] & [https://parts.igem.org/wiki/index.php?title=Part:BBa_K2170060 BBa_K2170060]]] | ||
This part contains a forbidden PstI site, which is generated by a point mutation in position 1.972. Because this restiction site makes the ligation of a BioBrick in the downstam region of this BioBrick by RFC[25] impossible the PstI site was removed by our team so that the resulting BioBrick [https://parts.igem.org/wiki/index.php?title=Part:BBa_K2170150 BBa_K2170150] matches the RFC[25] standard. | This part contains a forbidden PstI site, which is generated by a point mutation in position 1.972. Because this restiction site makes the ligation of a BioBrick in the downstam region of this BioBrick by RFC[25] impossible the PstI site was removed by our team so that the resulting BioBrick [https://parts.igem.org/wiki/index.php?title=Part:BBa_K2170150 BBa_K2170150] matches the RFC[25] standard. | ||
Revision as of 11:12, 16 October 2016
CMV promoter_mouse guanylate kinase_thymidine kinase 30
Additional characterization of BBaI712009 by [http://2016.igem.org/Team:LMU-TUM_Munich Munich 2016]:
This part contains a forbidden PstI site, which is generated by a point mutation in position 1.972. Because this restiction site makes the ligation of a BioBrick in the downstam region of this BioBrick by RFC[25] impossible the PstI site was removed by our team so that the resulting BioBrick BBa_K2170150 matches the RFC[25] standard.
Additionally the BioBrick BBa_K2170060 was assembled. This BioBrick contains a poly adenylation site, which was added for improved mRNA stability.
Introduction
Suicide gene therapy or gene-directed enzyme prodrug therapy (GDEPT) is widely used in cancer treatment. One of the most studied GDEPT systems is the herpes simplex virus thymidine kinase (HSV-TK) with purine nucleoside analog ganciclovir (GCV) as a prodrug. Systemic administration of the prodrug ganciclovir induces apoptosis only in cells transfected with HSV-thymidine kinase while the untransfected cells survive. Unlike human thymidine kinase, HSV-thymidine kinase is able to phosphorylate ganciclovir to form ganciclovir-monophosphate, which is then phosphorylated to ganciclovir-diphosphate and then to ganciclovir-triphosphate. Ganciclovir-triphosphate is then incorporated into DNA, which causes inhibition of DNA synthesis and subsequently leads to apoptosis (Ardiani et al., 2010).
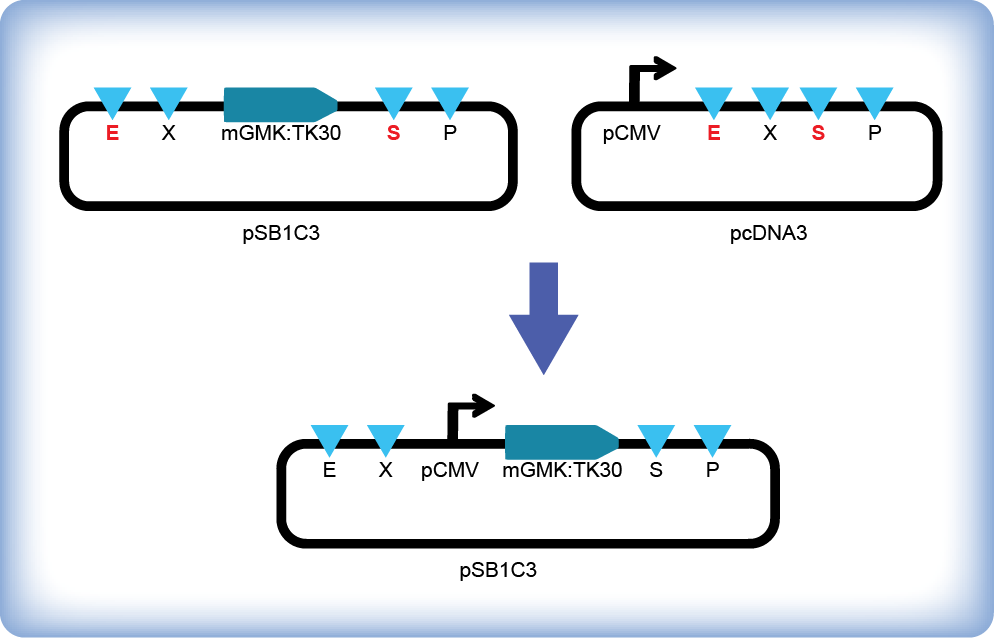
Following the BioBrick Standard Assembly technique iGEM team Slovenia inserted the mouse guanylate kinase – thymidine kinase fusion gene (mGMK:TK30, (BBa_K404113)) under the control of CMV promoter (Figure 1). HSV-TK mutant (TK30) contains six amino acid substitutions and shows enhanced sensitivity to ganciclovir (Kokoris et al., 1999), while mouse guanylate kinase improves phosphorylation of ganciclovir-monophosphate to ganciclovir-diphosphate, thus preventing accumulation of ineffective intermediate products (i.e. GCV-MP, GCV-DP) due to limited ability of endogenous guanylate kinase (Ardiani et al., 2010). We transfected HEK293 cells with the pPCMV-mGMK:TK30 plasmid and monitored cell survival after the addition of gancyclovir. We observed cells under an optical microscope daily and were attentive to any morphological signs of decreased viability. Additionally we stained cells with Hoechst and SytoxGreen514 dye to distinguish between live and dead cells with the help of a confocal microscope. We quantified viable cell numbers at different time points by cell counting and flow cytometry.
Figure 1: Assembly of mGMK:TK30 (BBa_K404113) under the control of CMV promoter. Insert (mGMK:TK30, (BBa_K404113)) was digested with EcoRI and SpeI and pcDNA3 vector with EcoRI and XbaI, loaded on an agarose gel and the appropriate bands were purified. Vector and insert were ligated and transformed into E. coli strain DH5α. The DNA sequence of the resulting plasmid pPCMV-mGMK_TK30 was confirmed by DNA sequencing.
Characterization
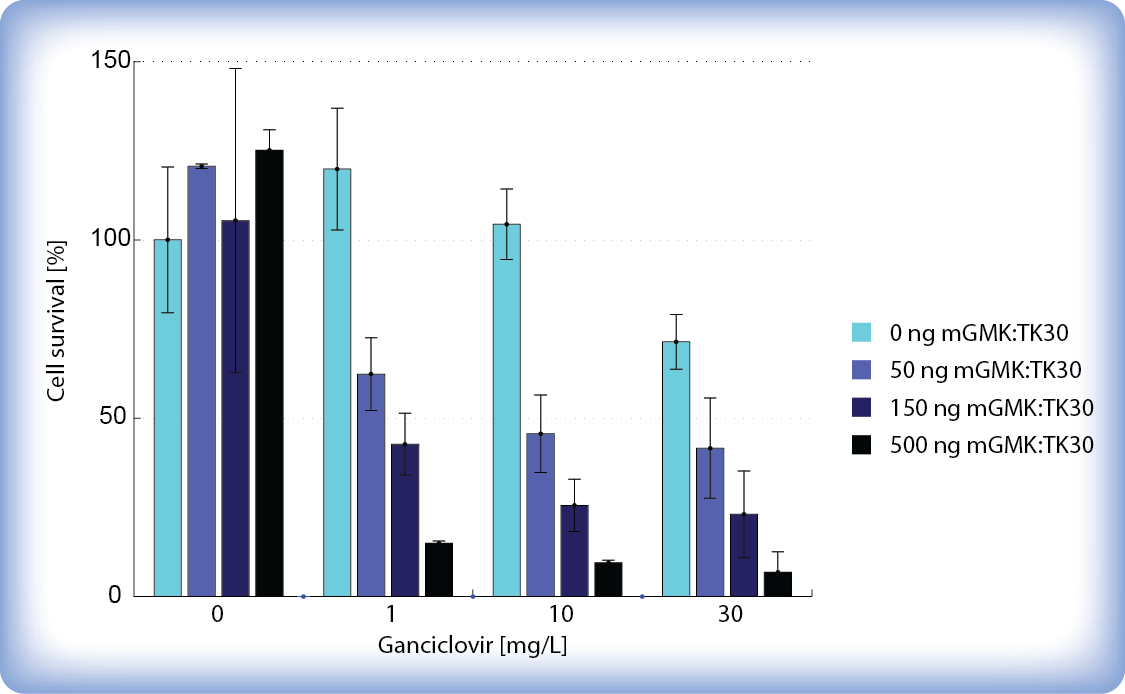
We tested different amounts of transfected pPCMV-mGMK:TK30 DNA, different concentrations of ganciclovir and different time periods after addition of ganciclovir to find the optimal conditions where our pCMV-mGMK:TK30-transfected cells would be most efficiently killed while untransfected cells would be left unharmed. We monitored pPCMV-mGMK:TK30-transfected ganciclovir-treated cells under a light microscope and discovered that as little as 40 hours of incubation with higher concentrations of ganciclovir (>30 μg/ml) is enough to kill over 50 % of our pPCMV-mGMK:TK30-transfected cells. After 65 hours of incubation with ganciclovir there was a decrease in cell number and survival already at low concentration of ganciclovir (1 μg/ml). Interestingly we also observed that higher amounts of transfected mGMK_TK30 influences cell viability even without ganciclovir. Also higher concentrations of ganciclovir became cytotoxic after longer periods of incubation. But more importantly, we showed that the HSV-TK/GCV system successfully kills cells even at low amounts of transfected mGMK:TK30 and upon addition of relatively low concentrations of ganciclovir (5 μg/ml).
We determined the number of viable cells after ganciclovir treatment by staining the cells with Trypan Blue (to distinguish live cells from the dead) and counting them. After 6 days of incubation with ganciclovir, the cytotoxic effect could be seen when only 1 μg/ml ganciclovir was added to pPCMV-mGMK:TK30 transfected cells while untransfected cells stayed unharmed. The number of viable cells decreased to <10 % when 10 μg/ml ganciclovir was added. These results demonstrate that our safety mechanism is effective even at low doses of ganciclovir, when only therapeutic cells are killed while untransfected cells are left unharmed.
Figure 2: Effect of ganciclovir (GCV) on HEK293 cell survival after 6 days of incubation with GCV.
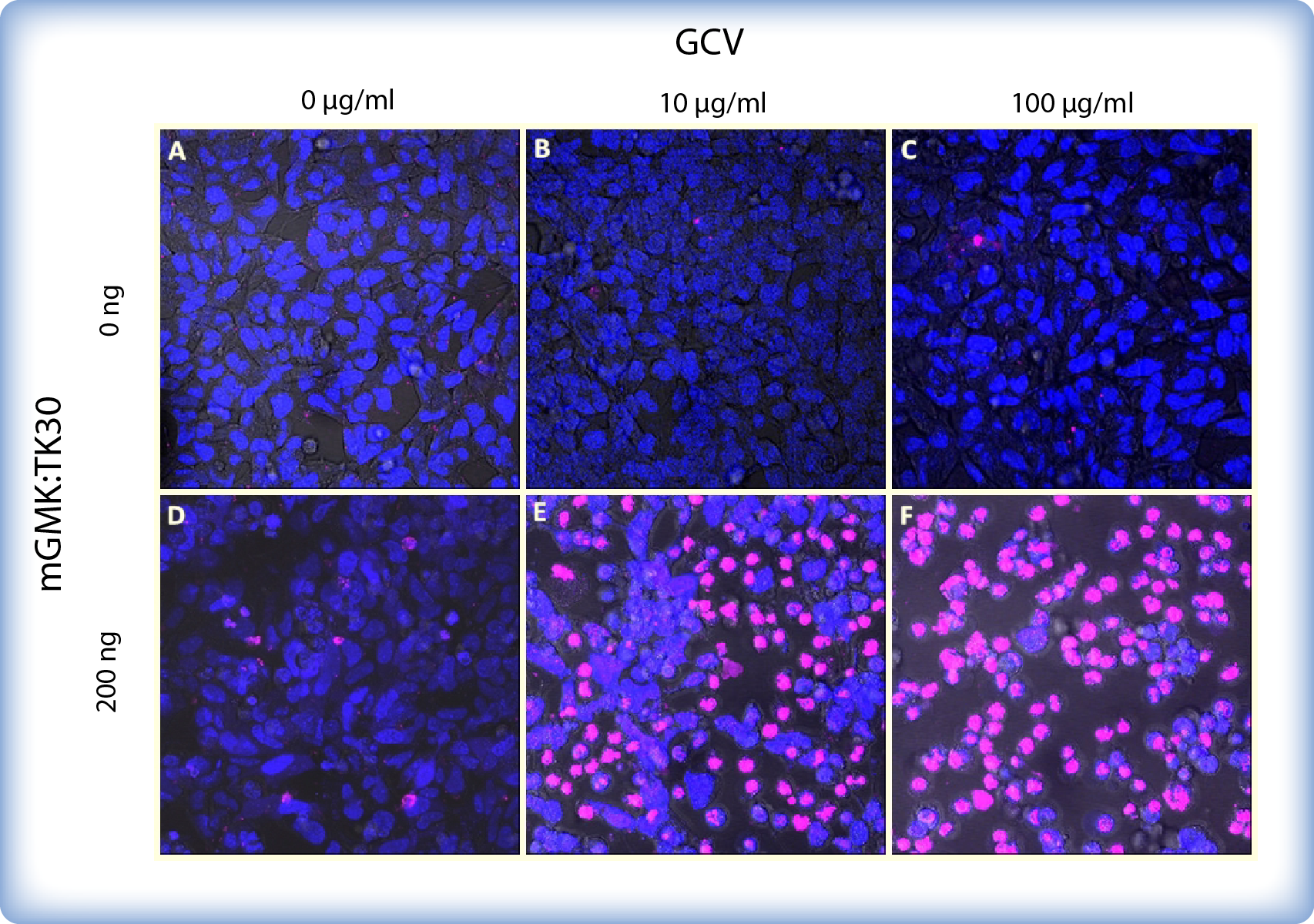
We also observed pPCMV-mGMK:TK30 transfected and ganciclovir treated HEK293 cells under a confocal microscope. Cells were transfected in a 12-well plate and stained with Hoechst and SytoxGreen514 dye, which differentially stain live and dead cells (Figure 3). Cell permeable Hoechst (depicted blue) binds to all nucleic acids and therefore stains all cells in a culture. On the other hand cell impermeable SytoxGreen (depicted pink) cannot cross the membrane of live cells and therefore stains only dead cells.
The cytotoxic effect of ganciclovir is evident at concentrations as low as 10 μg/ml and only a few viable cells where present upon addition of 100 μg/ml ganciclovir. On the other hand untransfected cells are not affected by ganciclovir. Moreover, transfection with pPCMV-mGMK:TK30 had no effect on cell survival if no ganciclovir was added.
Figure 3: HEK293 cells with or without mGMK_TK30 after 5 days of incubation with ganciclovir (GCV). Cells were transfected in a 8-well chamber and stained with Hoechst (blue) and SytoxGreen514 dye (dead/pink). (A-C) Untransfected cells: without GCV (A) or with 10 μg/ml (B) or 100 μg/ml (C) GCV show a high survival rate. (D) Cells transfected with 200ng pPCMV-mGMK:TK30 and without GCV show high cell viability. (E-F) Cells transfected with 200ng pPCMV_mGMK:TK30 and incubated with 10 μg/ml (E) or 100 μg/ml (F) GCV exhibit dramatically lower viability.
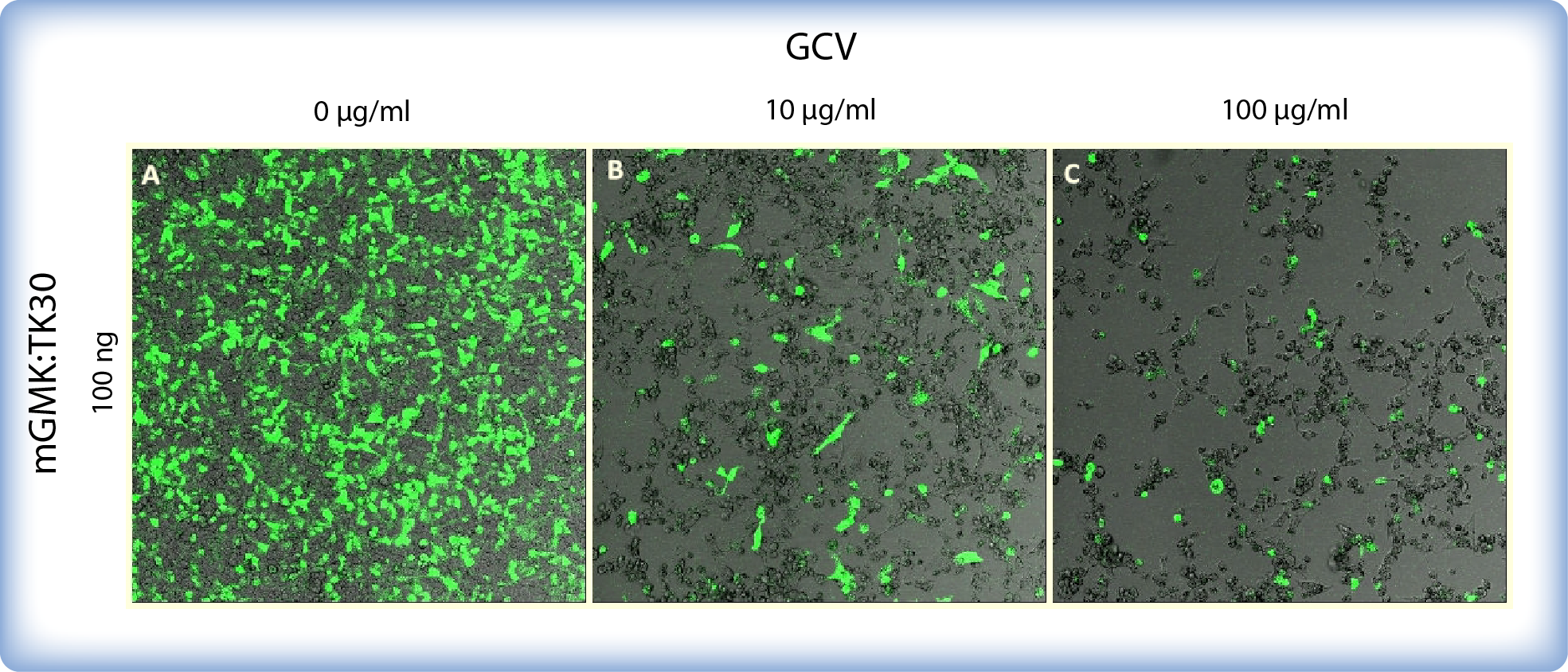
The bystander effect for mGMK_TK30 was demonstrated by Ardiani et al. (2010) who showed substantial ganciclovir-induced cell death although only a fraction of cells were transfected. There are several different mechanisms of bystander effect: the transfer of toxic metabolite ganciclovir-triphosphate through gap junctions (Elshami et al., 1996), endocytosis of apoptotic bodies generated from HSV-TK expressing cells by neighbouring untransfected cells (Freeman et al., 1993) and release of soluble factors (Drake et al., 2000). We also demonstrated that even if some cells do not express mGMK:TK30 (GFP was used as a control of transfection), ganciclovir affects practically all cells in a culture (Figure 4). If transiantly transfected therapeutic cells would be microencapsulated and used for therapy, the bystander effect ensures that all cells in a microcapsule would be killed upon addition of ganciclovir even if some of them would not express HSV-thymidine kinase.
Figure 4: Images of mGMK:TK30 and mCitrine transfected HEK293 cells incubated with ganciclovir. Cells were incubated with GCV for 5 days. Cells were transfected in a 8-well chamber with 100 ng pPCMV-mGMK:TK30 and 20 ng GFP. Cells depicted on (A) were not treated with GCV. Image (B) depicts cell death after incubation with 10 μg/mL GCV and image (C) shows drastic decrease in cell survival after incubation with 100 μg/mL GCV.
Trypan Blue and SytoxGreen514 only stain cells that are already dead. We also wanted to determine the number of cells that are still in the early stages of apoptosis. To do that we stained the cells with Annexin V and analyzed them with flow cytometry. Annexin V binds phosphatidylserine, which is located on the inner side of the plasma membrane. When apoptosis is induced phosphatidylserine is translocated to the extracellular side of the plasma membrane which enables staining with Annexin V.
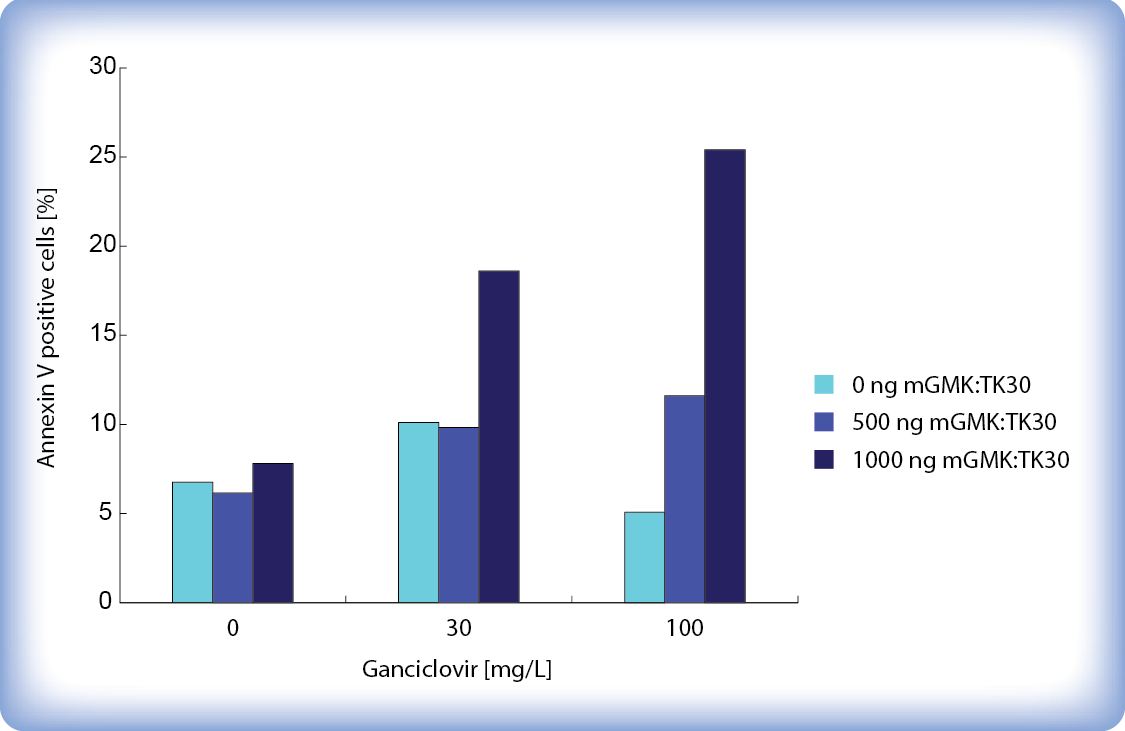
Figure 5 shows percent of Annexin V positive cells 65 hours after ganciclovir addition. The percent of apoptotic cells increases with higher ganciclovir concentrations and higher amounts of transfected pPCMV-mGMK:TK30. Transfection took place in a 12-well plate. Even though only around 25 % of cells stain with Annexin V we must keep in mind that after 65 hours ganciclovir already killed a large amount of cells which were therefore not included in the analysis.
Refrences
Ardiani, A., Sanchez-bonilla, M., and Black, M.E. (2010) Fusion Enzymes Containing HSV-1 Thymidine Kinase Mutants and Guanylate Kinase Enhance Prodrug Sensitivity In Vitro and In Vivo. Cancer 17, 86-96.
Drake, R.R., Pitlyk, K., McMasters, R.A., Mercer, K.E., Young, H., Moyer, M.P. (2000) Connexin-independent ganciclovir-mediated killing conferred on bystander effect-resistant cell lines by a herpes simplex virus- thymidine kinase-expressing colon cell line. Molecular therapy: the journal of the American Society of Gene Ther. 2, 515-523.
Elshami, A.A., Saavedra, A., Zhang, H., Kucharczuk, J.C., Spray, D.C., Fishman, G.I., Amin, K.M., Kaiser, L.R., Albelda, S.M. (1996) Gap junctions play a role in the ‘bystander effect’ of the herpes simplex virus thymidine kinase/ganciclovir system in vitro. Gene Ther. 3, 85-92.
Freeman, S.M., Abboud, C.N., Whartenby, K.A., Packman, C.H., Koeplin, D.S., Moolten, F.L., Abraham, G.N. (1993) The Bystander Effect : Tumor Regression When a Fraction of the Tumor Mass Is Genetically Modified. In Vitro 53, 5274-5283.
Kokoris, M.S., Sabo P., Adman E.T., Black, M.E. (1999) Enhancement of tumor ablation by a selected HSV-1 thymidine kinase mutant. Gene Ther. 6, 1415-1426.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1315
Illegal NgoMIV site found at 2220 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 707