Difference between revisions of "Part:BBa K2113004"
Saishreyas g (Talk | contribs) |
Saishreyas g (Talk | contribs) |
||
| Line 148: | Line 148: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | |||
| + | |||
| + | <!-- --> | ||
| + | These enzymes do not cut the peptide | ||
| + | |||
| + | <br>Asp-N endopeptidase | ||
| + | <br>Asp-N endopeptidase + N-terminal Glu | ||
| + | <br>Caspase1 | ||
| + | <br>Caspase10 | ||
| + | <br>Caspase2 | ||
| + | <br>Caspase3 | ||
| + | <br>Caspase4 | ||
| + | <br>Caspase5 | ||
| + | <br>Caspase6 | ||
| + | <br>Caspase7 | ||
| + | <br>Caspase8 | ||
| + | <br>Caspase9 | ||
| + | <br>Enterokinase | ||
| + | <br>Factor Xa | ||
| + | <br>Formic acid | ||
| + | <br>Glutamyl endopeptidase | ||
| + | <br>GranzymeB | ||
| + | <br>Hydroxylamine | ||
| + | <br>LysC | ||
| + | <br>LysN | ||
| + | <br>NTCB (2-nitro-5-thiocyanobenzoic acid) | ||
| + | <br>Pepsin (pH1.3) | ||
| + | <br>Proline-endopeptidase | ||
| + | <br>Staphylococcal peptidase I | ||
| + | <br>Thermolysin | ||
| + | <br>Thrombin | ||
| + | <br>Tobacco etch virus protease | ||
Revision as of 07:35, 14 October 2016
AMP without glycine
It consists of cationic anti-microbial peptides (AMP) with consecutive sequences of arginine and tryptophan (WRWRWR).This can be used as an antimicrobial agent against a wide range of gram positive and gram negative bacteria.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Mobyle@RPBS 3D Structure Prediction
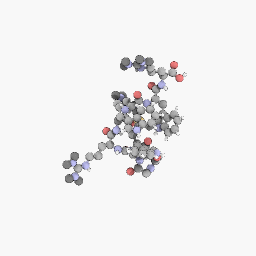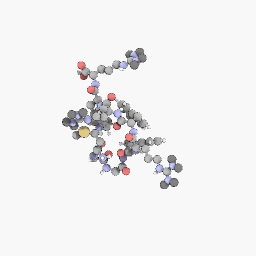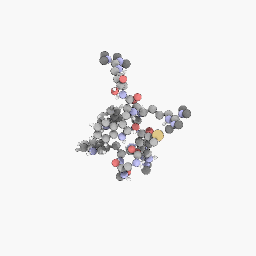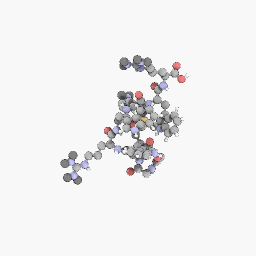
3D Structure
ProtParam Results
Number of amino acids: 7 Molecular weight: 1176.41 Theoretical pI: 12.30 Total number of negatively charged residues (Asp + Glu): 0 Total number of positively charged residues (Arg + Lys): 3
Atomic composition:
Carbon (C) 56
Hydrogen (H) 77
Nitrogen (N) 19
Oxygen (O) 8
Sulfur (S) 1
Formula: C56H77N19O8S1
Total number of atoms: 161
Extinction coefficients:
Extinction coefficients are in units of M-1 cm-1, at 280 nm measured in water.
Ext. coefficient: 16500
Abs 0.1% (=1 g/l): 14.026
Estimated half-life:
The N-terminal of the sequence considered is M (Met).
The estimated half-life is: 30 hours (mammalian reticulocytes, in vitro).
>20 hours (yeast, in vivo).
>10 hours (Escherichia coli, in vivo).
Instability index:
The instability index (II) is computed to be 172.23
This classifies the protein as unstable.
Aliphatic index: 0.00
Grand average of hydropathicity (GRAVY): -2.043
ExPASy Peptide Cutter Results
| Name of enzyme | No. of cleavages | Positions of cleavage sites |
|---|---|---|
| Arg-C proteinase | 3 | 3 5 7 |
| BNPS-Skatole | 3 | 2 4 6 |
| CNBr | 1 | 1 |
| Chymotrypsin-high specificity (C-term to [FYW], not before P) | 3 | 2 4 6 |
| Chymotrypsin-low specificity (C-term to [FYWML], not before P) | 4 | 1 2 4 6 |
| Clostripain | 3 | 3 5 7 |
| Iodosobenzoic acid | 3 | 2 4 6 |
| Pepsin (pH>2) | 4 | 1 2 4 6 |
| Proteinase K | 3 | 2 4 6 |
| Trypsin | 3 | 3 5 7 |
These enzymes do not cut the peptide
Asp-N endopeptidase
Asp-N endopeptidase + N-terminal Glu
Caspase1
Caspase10
Caspase2
Caspase3
Caspase4
Caspase5
Caspase6
Caspase7
Caspase8
Caspase9
Enterokinase
Factor Xa
Formic acid
Glutamyl endopeptidase
GranzymeB
Hydroxylamine
LysC
LysN
NTCB (2-nitro-5-thiocyanobenzoic acid)
Pepsin (pH1.3)
Proline-endopeptidase
Staphylococcal peptidase I
Thermolysin
Thrombin
Tobacco etch virus protease