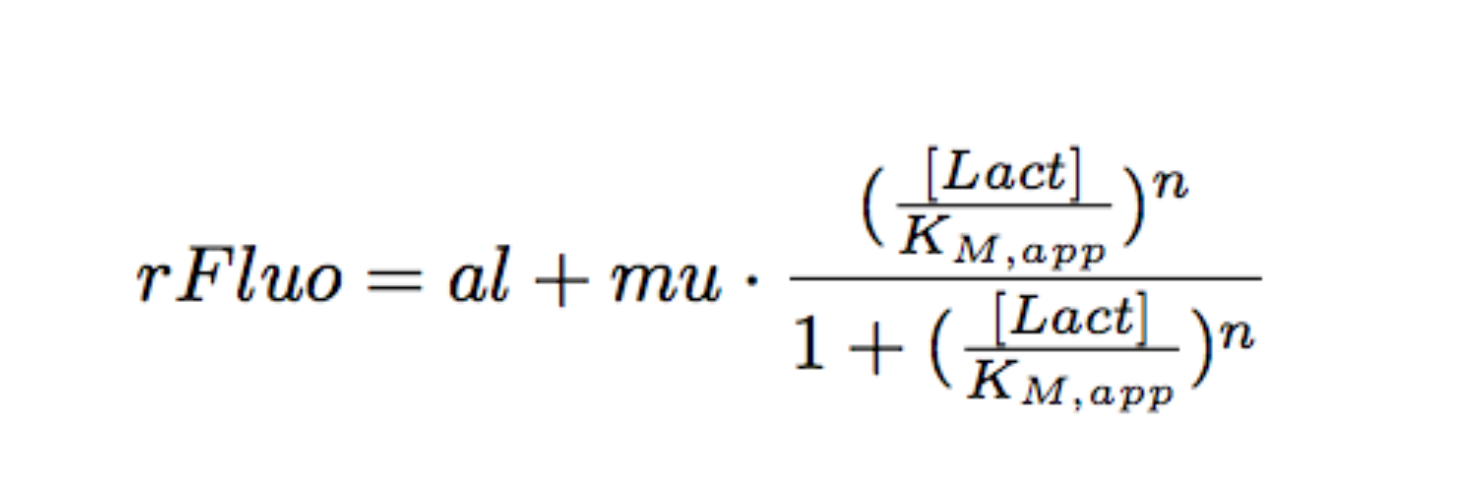Difference between revisions of "Part:BBa K1847008"
Phuongnguyen (Talk | contribs) |
Phuongnguyen (Talk | contribs) |
||
| Line 141: | Line 141: | ||
|rmse: 11.5851||rmse: 1.5452e+03||rmse: 150.3044||rmse: 803.1891 | |rmse: 11.5851||rmse: 1.5452e+03||rmse: 150.3044||rmse: 803.1891 | ||
|} | |} | ||
| − | + | <br> | |
<h1>Characterization and improvement of lldRO1-J23117-lldRO2 in D-Alanine auxotrophic bacteria</h1> | <h1>Characterization and improvement of lldRO1-J23117-lldRO2 in D-Alanine auxotrophic bacteria</h1> | ||
Revision as of 18:50, 13 October 2016
lldRO1-J23117-lldRO2
Collection of promoters regulated by LldR.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 78
Illegal NheI site found at 101 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
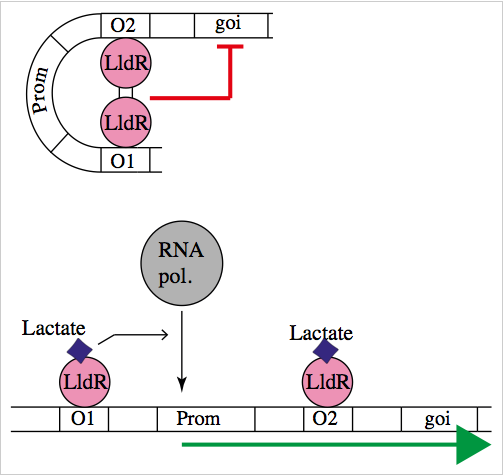
The natural promoter of LldR (Part:BBa_K822000) consists of two operators (O1 and O2) and a promoter which is intercalated between the operators. It regulates the expression of the lldPRD operon, and it is involved in L-lactate metabolism. This promoter is repressed by a dimer of LldR, possibly by forming a DNA loop that does not allow the RNA polymerase to bind to the promoter. LldR can also have a function as an activator [1]. The repression of the promoter can be removed by lactate.
Characterization of the promoter
We wanted to know how the promoter would respond to the lactate presence and how important is the architecture of the promoter in this response. For this regard, we built several promoters. Part:BBa_K1847002, Part:BBa_K1847003, Part:BBa_K1847004, Part:BBa_K1847005, Part:BBa_K1847006, Part:BBa_K1847007, Part:BBa_K1847008, and Part:BBa_K1847009.
Genetic design for characterization with LldR
To test our promoter we designed a plasmid containing the promoter with sfGFP in a medium copy plasmid pSEVA261 and transformed it into Escherichia coli TOP10. Then, we added a second plasmid containing lldR with a medium strong promoter (Part:BBa_J23118) and a strong RBS (Part:BBa_B0034) in pSEVA371 backbone. According to literature [1], LldR will repress the promoter and by the addition of lactate the production of GFP will start, as the repressor will be removed.
Genetic design for characterization with LldP-LldP
The second step for our characterization was to add an L-lactate permease (LldP), which despite its name is an active transporter of L-lactate, D-lactate and glycolate. To understand the effect of LldP (L-lactatep permease) in our system, we tried the following conditions:
- Medium copy plasmid with the promoter and sfGFP+ J23114-B0032-lldP-lldR (LldP and LldR are under the control of the same weak promoter) in low copy number plasmid
- Medium copy plasmid with the promoter and sfGFP + J23118-B0032-lldP-lldR (LldP and LldR are under the control of the same medium promoter) in low copy number plasmid
Characterization in a different medium
DMEM and RPMI mediums for mammalian cells where used to characterize this promoter. Two conditions were tested:
- Plasmid with the promoter and sfGFP + J23118-B0032-lldR
- Plasmid with the promoter and sfGFP+ J23118-B0034-lldP-lldR
Plate reader
E. coli TOP10 strains were grown overnight in Lysogeny Broth (LB) containing kanamycin (50 µg/mL) and chloramphenicol (12.5 µg/mL) at 37°C and 200 rpm. Cultures were diluted 1:50 in fresh LB with the corresponding antibiotic and transferred to a 96-well plate (200 µL/well). Cultures were grown for 90 min to arrive to exponential phase and then different concentrations of lactate were added. Samples were always made in triplicates and a blank of LB with the corresponding lactate concentration was done. During 7 h the absorbance at OD600 and fluorescence (excitation 488 nm and emission 530 nm) were measured with intervals of 7 min. The plate was always kept at 37°C in a Tecan Infinite M200 Pro Plate Reader. We calculated dose-response curves from the exponential phase of the bacteria after normalizing fluorescence by optical density.
Modeling
Each experimental data set was fitted to a Hill function using the Least Absolute Residual method (Fitting Toolbox in MatLab).
Where:
- al: leakiness in µM/min
- mu: constant in µM/min
- KM: half-maximum effective concentration (a representation of sensitivity) in µM
- n: Hill coefficient (a representation of cooperativity), unitless
| J23118-B0034-lldR | J23118-B0034-lldP-lldR | J23114-B0032-lldP-lldR | |
|---|---|---|---|
| Coefficient values | KM=1075 (889.5, 1261) | KM=1751 (918.5, 2584) | KM=337.7 (152.4, 522.9) |
| al = 479.3 (-142.2, 1101) | al = 550 (-1635, 2735) | al = 3444 (2571, 4317) | |
| mu = 7311 (6583, 8039) | mu = 1.318e+04 (1.03e+04, 1.607e+04) | mu = 4886 (3817, 5954) | |
| n1 = 1.326 (1.063, 1.588) | n1 = 0.9388 (0.5614, 1.316) | n1 = 1.09 (0.531, 1.649) | |
| Goodness to fit | sse: 4.2545e+07 | sse: 6.4285e+08 | sse: 2.7027e+08 |
| rsquare: 0.9964 | rsquare: 0.9879 | rsquare: 0.9743 | |
| fe: 7 | dfe: 7 | dfe: 7 | |
| adjrsquare: 0.9949 | adjrsquare: 0.9828 | adjrsquare: 0.9633 | |
| rmse: 2.4653e+03 | rmse: 9.5831e+03 | rmse: 6.2137e+03 |
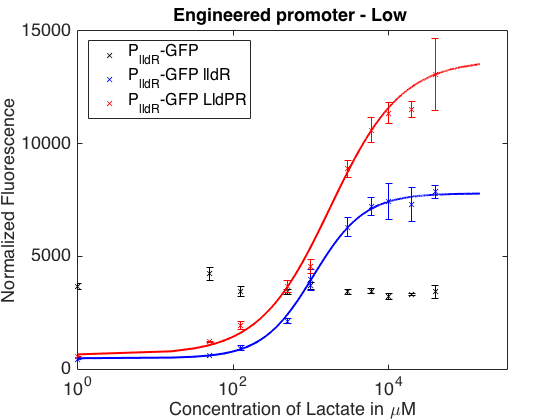
As can be seen in the picture above, when PlldR GFP is alone, the fluorescence levels do not vary with lactate concentration. However, if PlldR GFP is expressed in a system with constitutive expression of LldR, at low concentrations of lactate there is repression of the promoter, while at high concentrations there is activation. Activation is higher when there is constitutive expression of LldP-LldR. KM is shifted regarding the wild-type promoter, which makes this promoter more sensitive.
The promoter has higher ON/OFF ratios than the wild-type promoter (see Table 1). Its KM is higher than the one from the wild-type promoter, meaning that is has lower affinity for the substrate.
This promoter is one from our designed and tested synthetic promoters based on the wild-type PlldR. In the following table, one can see the ON/OFF ratios and the KM of the wild-type promoter and the three more prominent members of the library.
Table 1: ON/OFF ratio
| LldR | Additional LldP | |||
|---|---|---|---|---|
| J23118-B0034-lldR | J23118-B0034-lldP-lldR | J23114-B0032-lldP-lldR | ||
| Part:BBa_K822000 (wild-type) | 10.35 | 8.04 | 1.16 | |
| Part:BBa_K1847008 | 15.26 | 23.96 | 1.42 | Increasing promoter strength |
| Part:BBa_K1847009 | 2.5 | 24.34 | 0.96 | |
| Part:BBa_K1847007 | 2.15 | 3.85 | 1.29 | |
Table 2: KM (µM)
| J23118-B0034-lldR | J23118-B0034-lldP-lldR | J23114-B0032-lldP-lldR | |
|---|---|---|---|
| Part:BBa_K822000 (wild-type) | 955 | 1930 | 720.2 |
| Part:BBa_K1847008 | 1075 | 1751 | 337.5 |
| Part:BBa_K1847009 | 977.5 | 2361 | 459.8 |
| Part:BBa_K1847007 | 697.7 | 1977 | 1337 |
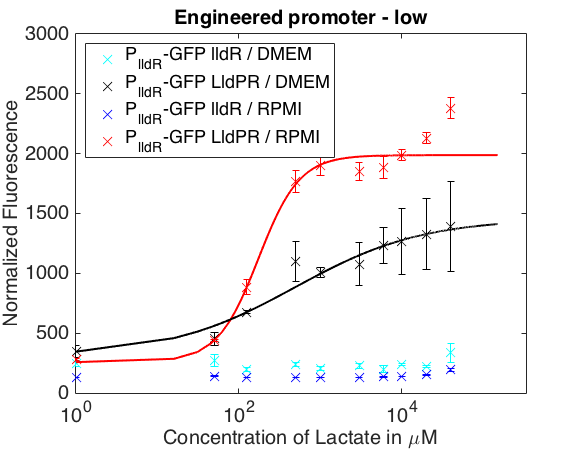
The graph below shows the behavior of our promoter in two mediums usually used for mammalian cell cultures. The promoter does not respond to lactate concentrations in DMEM while in RPMI it shows a behavior similar to the one in LB.
| RPMI | DMEM | |||
|---|---|---|---|---|
| J23118-B0034-lldR | J23118-B0034-lldP-lldR | J23118-B0034-lldR | J23118-B0034-lldP-lldR | |
| Coefficient values | K=3.389e+04 (-1.7e+05, 2.378e+05) | K=173.6 (15.19, 331.9) | K=5e+04 (-2.563e+05, 3.563e+05) | K = 448.4 (52.45, 844.3) |
| al = 136.2 (130.8, 141.7) | al = 253 (-584.6, 1091) | al = 230.3 (202.7, 258) | al = 314.1 (138.6, 489.5) | |
| mu = 92.3 (-659.7, 844.3) | mu = 1735 (863.1, 2607) | mu = 253.2 (-1098, 1605) | mu = 1085 (907.3, 1263) | |
| n1 = 3.165 (-13.87, 20.2) | n1 = 1.7 (0.05844, 3.341) | n1 = 1.5 (fixed at bound) | n1 = 0.7 (fixed at bound) | |
| Goodness to fit | sse: 939.5028 | sse: 1.6713e+07 | sse: 1.8073e+05 | sse: 5.1609e+06 |
| rsquare: 0.9640 | rsquare: 0.8846 | rsquare: 0.7522 | rsquare: 0.9608 | |
| fe: 7 | dfe: 7 | dfe: 8 | dfe: 8 | |
| adjrsquare: 0.9485 | adjrsquare: 0.8351 | adjrsquare: 0.6902 | adjrsquare: 0.9510 | |
| rmse: 11.5851 | rmse: 1.5452e+03 | rmse: 150.3044 | rmse: 803.1891 | |
Characterization and improvement of lldRO1-J23117-lldRO2 in D-Alanine auxotrophic bacteria
Group: Team NUS_Singapore 2016
Authors: Keshiniy Madivannan, Janice Darikho and Nguyen Hoang Diem Phuong
Introduction
Due to the ability to deregulate normal cellular energetics, cancer cells are found to undergo aerobic glycolysis followed by lactic acid fermentation in the cytosol, resulting in the elevated production and accumulation of L-lactate in the surrounding environment (Chen et al., 2007; Vander et al., 2009; Alfarouk et al., 2015). The RIOT sensor collection was designed to detect increased level of L-lactate produced by tumor cells in the microenviroment. A lactate sensitive promoter obtained from 2015 ETH Zurich team was used as starting material. Our designed sensor has to be extremely sensitive to small changes in lactate concentration. This means that our sensor needs to have no basal expression and significant increase in reporter output (Alanine Racemase, ALR) in the presence of lactate. We minimise the basal expression of our reporter by linking different strength RBS parts with LldPR promoter (BBa_K822000) or the engineered promoter (BBa_K1847008). Our strategy allows strict amount of reporter expression, thus increasing sensitivity of detection, and minimise false positive result. ALR is required for synthesis of D-alanine, an essential amino acid for bacterial survival (Walsh, 1989). Bacteria transformed with an ideal sensor are expected to survive in mild lactate concentration of the blood. Once reaching tumour site, their growth will be increased by the high lactate concentration there which will enhance the detection signal or the delivery of biomolecules into tumour cells.
Methods
We transformed E. coli Nissle ∆alr, a D-alanine auxotrophic mutant bacteria with RIOT sensors which express ALR in the presence of L-lactate. ALR will rescue the transformed bacteria grown in media without D-alanine supplement. Therefore, the performance of RIOT sensors were assessed by the growth rate of transformed bacteria over a range of lactate concentration in M9 minimal media via natural logarithm of OD600.
Results
D-alanine auxotrophic mutant bacteria were transformed with plasmids containing ALR sequence under the control of lldRO1-J23117-lldRO2 or RIOT sensor 7. Their overnight culture were spun down and resuspended with M9 minimal media without D-alanine supplement. Then, they were subjected to different treatments (Fig 3):
- M9 without D-alanine supplement
- M9 with D-alanine supplement (50 µg/ml)
- M9 with addition of lactate of 10-4 to 10-2 M.
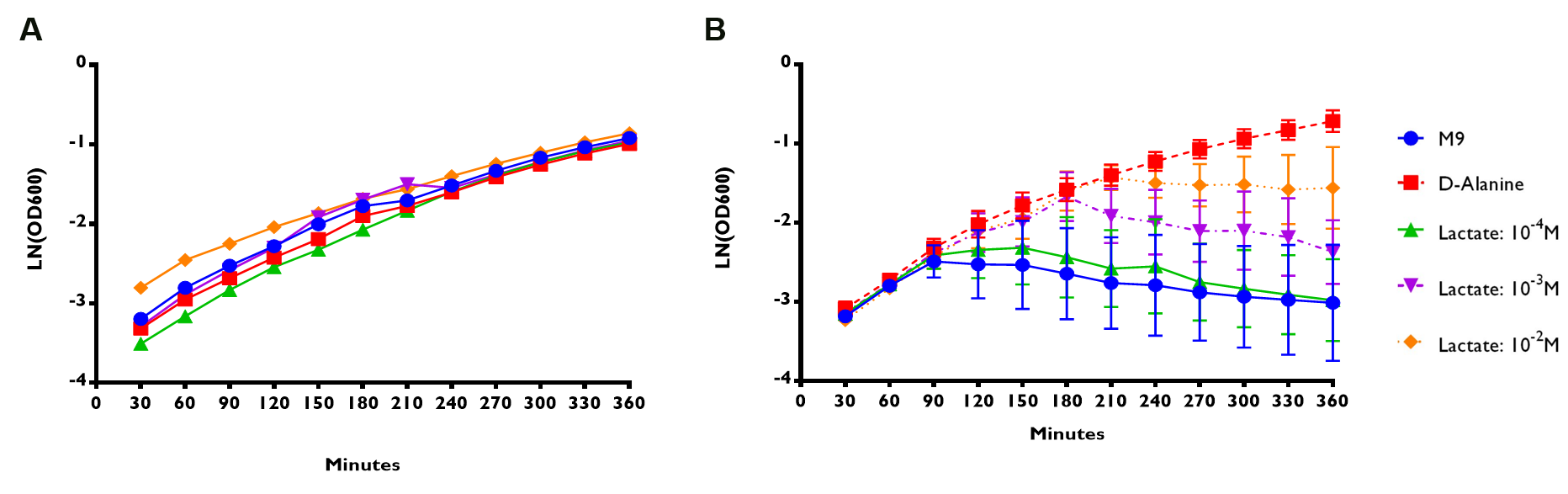
OD600 readings of transformed bacteria under different treatments were measured for 6 hours. LN of OD600 readings reflects the bacterial growth rate. (A) RIOT sensor 1: p70-34-ALR-Terminator (BBa_K1897019) consists of lldRO1-J23117-lldRO2 promoter ligated to Alanine Racemase (ALR). n=1 ± SEM. (B) RIOT sensor 7: p70-33-ALR-Terminator (BBa_K1897021) consists of our modified promoter ligated to ALR. n=3 ± SEM.
We used linear regression analysis to fit the data of bacterial growth rate using four data points of LN (OD600) values from 90 to 180 minutes of each sensor. This is because the growth rate of each constructs appeared to deviate from each other at time point 90 minute (Fig 3).
Table 4: Comparisons of bacteria growth rate between RIOT sensor 1 and 7
RIOT sensor 1: p70-34-ALR-Terminator (BBa_K1897019) consists of lldRO1-J23117-lldRO2 promoter ligated to Alanine Racemase (ALR). n=1 ± SEM.
RIOT sensor 7: p70-33-ALR-Terminator (BBa_K1897021) consists of our modified promoter ligated to ALR. n=3 ± SEM.
Doubling time – LN (2) / Growth rate
| RIOT sensor No. | Lactate concentration (M) | Growth rate (min-1) | Doubling time (min) | R square |
|---|---|---|---|---|
| 1: p70-34-ALR-Terminator | 10-4 | 0.00935±0.000594 | 74.3±4.73 | 0.992 |
| 10-3 | 0.0107±0.000505 | 64.9±3.06 | 0.996 | |
| 10-2 | 0.00659±0.000173 | 105.2±2.76 | 0.999 | |
| 7: p70-33-ALR-Terminator | 10-4 | No growth was observed | ||
| 10-3 | 0.00756±0.000789 | 92.7±9.67 | 0.979 | |
| 10-2 | 0.00880±0.000863 | 79.5±7.80 | 0.981 | |
Discussion and Conclusion
The results shown in Fig. 3 suggested that RIOT 1 with lldRO1-J23117-lldRO2 (BBa_K1847008) exhibited high leakiness. The transformed bacteria were able to survive after 6 hours even without addition of lactate. Moreover, table 4 showed that transformed bacteria with sensors 1 and 2 under different treatments appeared to have similar initial growth rate compared to those were grown in the media containing D-alanine supplement (0.008 – 0.009 min-1, P-value < 0.01). This is probably because the amount of ALR expressed at the lowest lactate concentration is still sufficient to convert enough L-alanine into D-alanine, which help the bacteria to maintain their normal growth. The insignificant difference in growth rate of bacteria under various lactate concentrations indicated that RIOT sensor using lldRO1-J23117-lldRO2 promoter is not good candidate to detect lactate in tumour microenvironment.
In contrast, RIOT sensors 7, p70-33-ALR-Terminator (BBa_K1897021) exhibited very minimal leakiness (Fig 3). The transformed bacteria were unable to grow in the absence or at low lactate concentration (10-4 M). The addition of lactate at high concentration (10-3 and 10-2 M) rescued the transformed bacteria grown in media without D-alanine. According to table 1, the growth rate of bacteria transformed with sensor 7 grown in lactate concentration of 10-4 M was not significantly different from 0. RIOT sensor 7 also showed an increase in growth rate or a decrease in doubling time from 92.7 to 79.5 minutes when lactate concentration increased from 10-3 M to 10-2 M. In conclusion, we have characterized the output of lldRO1-J23117-lldRO2 (BBa_K1847008) in a new chassis which was E. coli Nissle ∆alr, a D-alanine auxotrophic mutant bacteria to detect lactate in tumour microenvironment. We also improved the sensitivity of this promoter with our RIOT sensor 7 without the need of adding LldP and LldR. The result shows that we have successfully minimized the leakiness and enhance the sensitivity to lactate of lldRO1-J23117-lldRO2. Bacteria transformed with sensor 7 are expected to be able to survive in mild lactate concentration of the blood and once reaching tumour site, their growth will be enhanced by the high lactate concentration there which will enhance the detection signal or the delivery of biomolecules into tumour cells.
References
- J Bacteriol. 2008 Apr; 190(8): 2997–3005.