Difference between revisions of "Part:BBa K1694037"
| Line 6: | Line 6: | ||
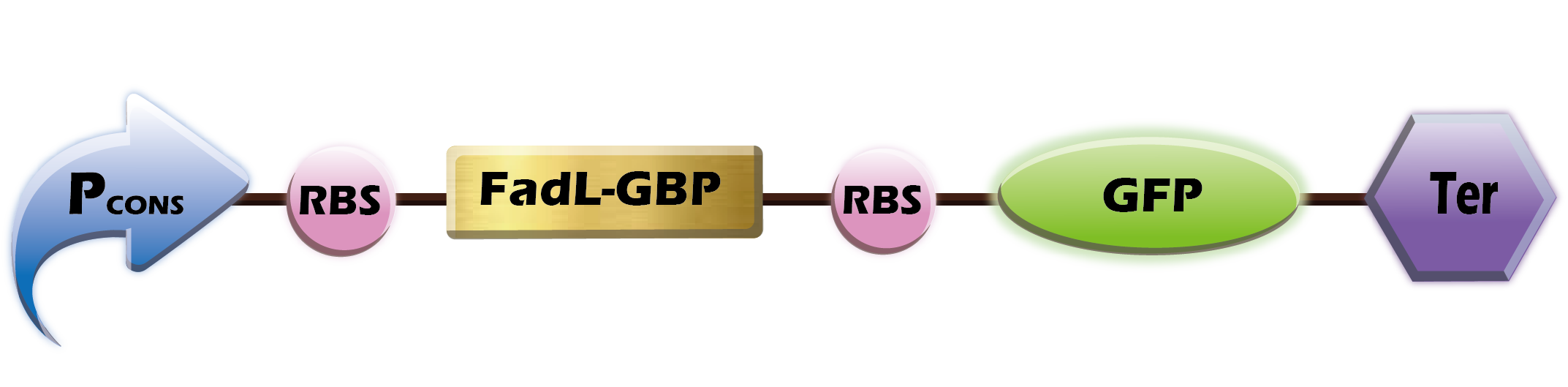
[[File:GBPGFP.png|600px|thumb|center|'''Fig.1''' P<sub>cons</sub>+B0034+FadL-GBP+B0030+GFP+Ter]] | [[File:GBPGFP.png|600px|thumb|center|'''Fig.1''' P<sub>cons</sub>+B0034+FadL-GBP+B0030+GFP+Ter]] | ||
| − | The transmembrane protein, FadL, fused with the GBP | + | The transmembrane protein, FadL, fused with the GBP, |
FadL-GBP, can act as an anchoring motif for displaying GBP on the surface of ''E. coli''. To achieve the convenient observation, the genetic sequence of green fluorescent protein was introduced in our design. With the dual display of the GBP and the GFP, the ''E. coli'' can not only bind on the gold surface but also produce green fluorescent signal.<br> | FadL-GBP, can act as an anchoring motif for displaying GBP on the surface of ''E. coli''. To achieve the convenient observation, the genetic sequence of green fluorescent protein was introduced in our design. With the dual display of the GBP and the GFP, the ''E. coli'' can not only bind on the gold surface but also produce green fluorescent signal.<br> | ||
Revision as of 07:30, 22 September 2015
Pcons+B0034+FadL-GBP+B0030+GFP+J61048
Introduction
The transmembrane protein, FadL, fused with the GBP,
FadL-GBP, can act as an anchoring motif for displaying GBP on the surface of E. coli. To achieve the convenient observation, the genetic sequence of green fluorescent protein was introduced in our design. With the dual display of the GBP and the GFP, the E. coli can not only bind on the gold surface but also produce green fluorescent signal.
Introduction of basic parts:
FadL-GBP
Experiment
After assembling the DNA sequences from the basic parts, we recombined each Pcons+B0034+FadL-GBP+B0030+GFP+J61048 gene to pSB1C3 backbones and conducted a PCR experiment to check the size of each of the parts. The DNA sequence length of these parts is around 2000~2200 bp. In this PCR experiment, the scFv products size should be near at 2200~2400 bp. The Fig. 2 showed the correct size of the scFv, and proved that we successful ligated the scFv sequence onto an ideal backbone.
We intended to make the observation of the result more directly by producing green fluorescence at the same time. To make the comparison of the binding efficiency, we selected the genetic sequence, Pcons + RBS +GFP +Ter, as the negative control group. Because there was no existence of fully-functioned phenomenon, there was no positive control in our experiment.
According to the pictures shown above, we can see that there are more E. coli with gold-binding polypeptide being observed on the gold chip than the E. coli without gold-binding polypeptide. It indicates that the E. coli with gold-binding polypeptide can attach on gold chip more effectively than others without gold-binding polypeptide.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 499
Illegal BamHI site found at 985 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 275
Illegal NgoMIV site found at 767
Illegal NgoMIV site found at 1192
Illegal NgoMIV site found at 2102
Illegal AgeI site found at 908 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 2018