Difference between revisions of "Part:BBa K1639004"
(→References) |
(→Characterization) |
||
| Line 21: | Line 21: | ||
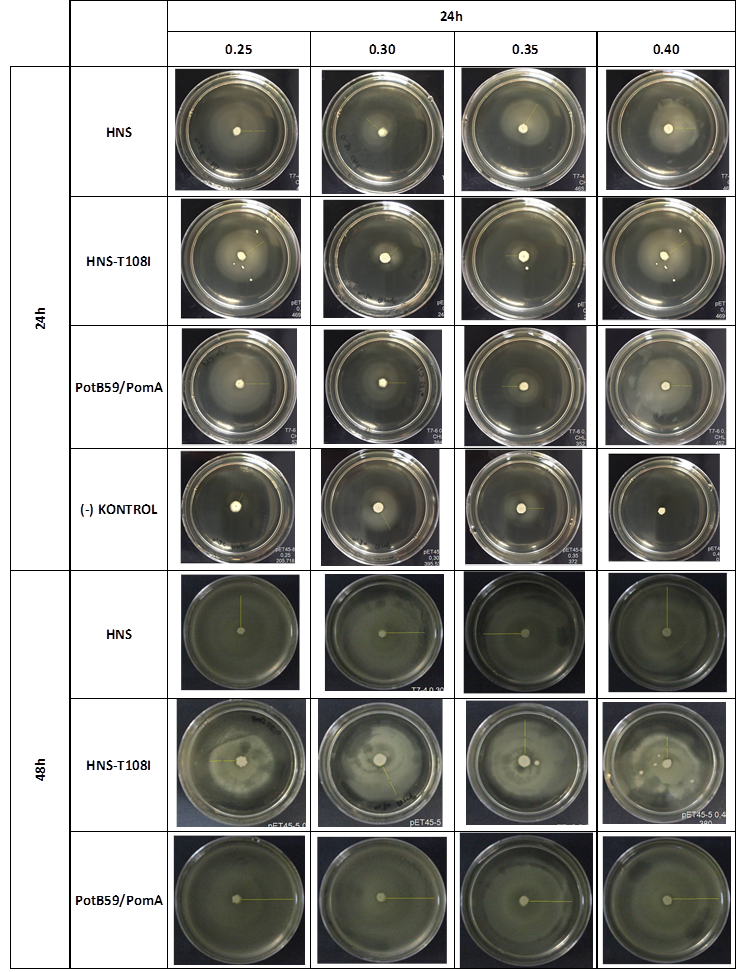
After showing that we produced required proteins successfully, in order to learn if they are functional, we did a motility assay called ‘plug assay’. We inoculated bacteria in different concentrations into the soft agars and incubated them at 37°C. After this, we measured their colony diameters at 24. and 48. hours. The results are shown below. | After showing that we produced required proteins successfully, in order to learn if they are functional, we did a motility assay called ‘plug assay’. We inoculated bacteria in different concentrations into the soft agars and incubated them at 37°C. After this, we measured their colony diameters at 24. and 48. hours. The results are shown below. | ||
| − | [[File:ATOMS-Turkiye_ulcer_incmot_3.9.png|center | + | [[File:ATOMS-Turkiye_ulcer_incmot_3.9.png|center|'''Figure 5''']] |
plasmid transformed bacteria''' HNS:pET45-b-HNS''' plasmid transformed bacteria '''HNS-T108I:pET45-b-HNS-T108I''' plasmid transformed bacteria''' PotB59/PomA :pET45-b-PotB59/PomA''' plasmid transformed bacteria.The measurements of 0.25 and 0.4 concentrations in first 24. hour period, gives us significant positive results . If we compare the results,colony diameters are ordered like this; (-) kontrol < Wild type HNS < PotB59/PomA. The results of 0.30 and 0.35 agar concentrations doesn’t give significant information. Besides, in 48. hour of incubation the diameter of colonies reached to plate’s diameter, so measurement results didn’t show any difference than 24. hour’s. | plasmid transformed bacteria''' HNS:pET45-b-HNS''' plasmid transformed bacteria '''HNS-T108I:pET45-b-HNS-T108I''' plasmid transformed bacteria''' PotB59/PomA :pET45-b-PotB59/PomA''' plasmid transformed bacteria.The measurements of 0.25 and 0.4 concentrations in first 24. hour period, gives us significant positive results . If we compare the results,colony diameters are ordered like this; (-) kontrol < Wild type HNS < PotB59/PomA. The results of 0.30 and 0.35 agar concentrations doesn’t give significant information. Besides, in 48. hour of incubation the diameter of colonies reached to plate’s diameter, so measurement results didn’t show any difference than 24. hour’s. | ||
Latest revision as of 09:00, 21 September 2015
H-NS
global DNA-binding transcriptional dual regulator H-NS
Usage and Biology
Many species of bacteria have flagellar motility that is achieved by rotating surface-exposed organelles The rotation of the flagella is controlled by a motor complex embedded in the inner membrane (reviewed in Refs. 2 and 3 The motor region of E. Coli’s flagella is composed of several parts. The stator complex, made up of the proteins MotA-MotB, has a transmembrane characteristic and ensures the generation of a proton channel (4-6). The rotor complex involves the interaction of three other proteins, FliG, FliM, and FliN(7,8). All three rotor proteins are involved in the processes of flagellar assembly, switching, and rotation (9,10). However, FliG is predominately involved in torque generation (8, 10, 11), whereas FliM mainly functions in switching rotor direction (12). The precise role of FliN is the least well defined, but it may participate in flagella protein-specific export and assembly (13,14)
HNS is a transcription factor that plays a role in the regulation of gene expression in E. Coli. It plays an activator role at the level of protein synthesis for many genes involved in bacterial motility. . . In many studies it was shown that HNS is essential for the synthesis of proteins that make up the rotor and stator. The flhCD operon is a positive regulator of HNS which is responsible for the synthesis of many proteins that composes the structure of the bacterial flagella. Studies show that knock-out of the gene that codes the HNS protein results in non-motile E. Coli.
Above, we mentioned three proteins that make up the stator portion of the bacterial flagella.. These proteins are FliG, FliM and FliN and these three proteins have different activities. FliM is a regulator protein that is responsible for determining in which direction the bacterial flagella will be oriented. Although the role of FliN is not precisely known, it is thought that it acts as a guide for the assembly of the flagella’s membrane/for attachment of flagella to the membrane. As for FliG, this protein is responsible for the rotation speed and torque power of the bacterial flagella. For our aims in this part of the project, which are to increase the speed and power of the bacterial flagella, we decided to focus on FliG.
In a study by Gina M. Donato and Thomas H. Kawula, it was shown that HNS is not only a transcription factor but also that it can change some protein’s activities through direct interactions with them. . In studies including fluorescence anistropy and chemical cross-linking, it was shown that there is a physical interaction between HNS and FliG proteins.
Characterization
SOFT AGAR PLUG ASSAY
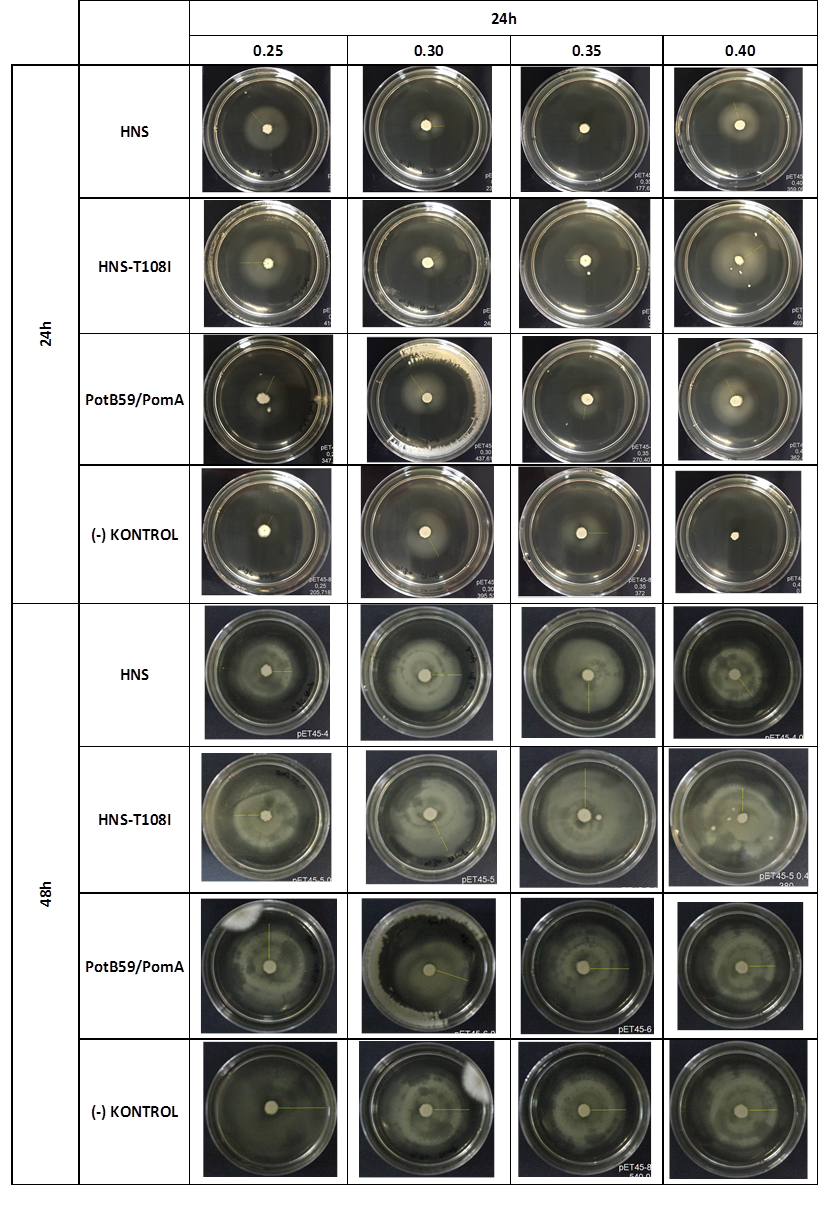
After showing that we produced required proteins successfully, in order to learn if they are functional, we did a motility assay called ‘plug assay’. We inoculated bacteria in different concentrations into the soft agars and incubated them at 37°C. After this, we measured their colony diameters at 24. and 48. hours. The results are shown below.
plasmid transformed bacteria HNS:pET45-b-HNS plasmid transformed bacteria HNS-T108I:pET45-b-HNS-T108I plasmid transformed bacteria PotB59/PomA :pET45-b-PotB59/PomA plasmid transformed bacteria.The measurements of 0.25 and 0.4 concentrations in first 24. hour period, gives us significant positive results . If we compare the results,colony diameters are ordered like this; (-) kontrol < Wild type HNS < PotB59/PomA. The results of 0.30 and 0.35 agar concentrations doesn’t give significant information. Besides, in 48. hour of incubation the diameter of colonies reached to plate’s diameter, so measurement results didn’t show any difference than 24. hour’s.
FUNCTIONAL ASSAY
We repeated functional assays after we cloned the genes into PSB1C3-T7. The results are shown below.
The measurements which made at first 24. hour period, gives us significant positive results . If we compare the results,colony diameters are ordered like this; (-) control < Wild type HNS < HNS-T108I< PotB59/PomA
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 2
Illegal XhoI site found at 426 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 162
References
[1]Gina M. Donatoand Thomas H. Kawula(1998)TheJournalBiologicalChemistryVol. 273, No. 37, 24030-24036
[2]Macnab, R. M. (1992) Annu. Rev. Genet. 26, 131–158
[3]Macnab, R. M. (1996) EscherichiacoliandSalmonellatyphimurium Cellular andMolecularBiology (Neidhardt, F. C.,Curtiss, R., III, Ingraham, J. L., Lin, E. C. C.,Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M.,andUmbarger, H. E., eds) 2nd Ed., pp. 123–145, American SocietyforMicrobiology, Washington, D. C.
[4]Blair, D. F.,andBerg, H. C. (1990) Cell 60, 439–449
[5]Blair, D. F.,andBerg, H. C. (1991) J. Mol. Biol. 221, 1433–1442
[6]Garza, A. G., Harris-Haller, L. W., Stoebner, R. A., andManson, M. D. (1995) Proc. Natl. Acad. Sci. U. S. A. 92, 1970–1974
[7]Marykwas, D. L.,Schmidt, S. A., andBerg, H. C. (1996) J. Mol. Biol. 256,564–576
[8]Tang, H.,Braun, T. F., andBlair, D. F. (1996) J. Mol. Biol. 261, 209–221
[9]Yamaguchi, S.,Fujita, H., Ishihara, A., Aizawa, S.-I., andMacnab, R. M. (1986) J. Bacteriol. 166, 187–193
[10]Irikura, V. M.,Kihara, M., Yamaguchi, S., Sockett, H., andMacnab, R. M. (1993) J. Bacteriol. 175, 802–810
[11]Lloyd, S. A.,Tang, H., Wang, X., Billings, S., andBlair, D. F. (1996) J. Bacteriol. 178, 223–231
[12]Welch, M.,Oosawa, K., Aizawa, S.-I., andEisenbach, M. (1993) Proc. Natl. Acad. Sci. U. S. A. 90, 8787–8791
[13]Vogler, A. P.,Homma, M., Irikura, V. M., andMacnab, R. M. (1991) J. Bacteriol. 173, 3564–3572
[14]Tang, H.,Billings, S., Wang, X., Sharp, L., andBlair, D. F. (1995) J. Bacteriol. 177, 3496–3503
[15]Asai, Y., T. Yakushi, I. Kawagishi, and M. Homma. 2003. Ion-couplingdeterminants of Na + -drivenand H + -drivenflagellarmotors. J. Mol. Biol. 327: 453-