Difference between revisions of "Part:BBa K1806004"
Abdulkadir (Talk | contribs) (→Cloning) |
|||
| Line 8: | Line 8: | ||
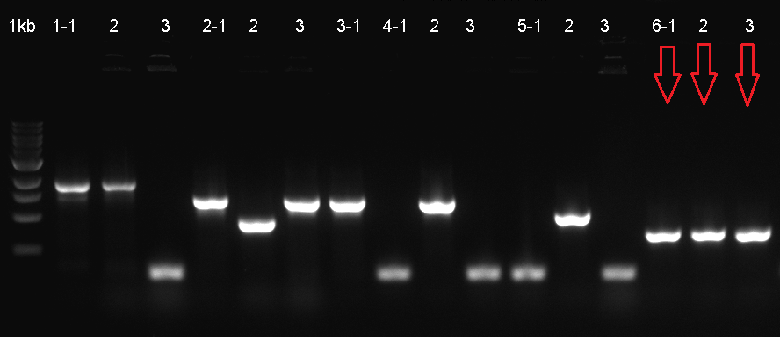
The composite system was ligated to the cloning vector psB1C3, to be transformated and then verified with Colony PCR. The verified ligation was then moved on to the digestion step to be cut with the EcoR1 and Pst1 restriction enzymes. The gel run of the cut parts were portrayed down below. The part was tagged as G-Block 6 and sample 6-2 yielded accurate results. | The composite system was ligated to the cloning vector psB1C3, to be transformated and then verified with Colony PCR. The verified ligation was then moved on to the digestion step to be cut with the EcoR1 and Pst1 restriction enzymes. The gel run of the cut parts were portrayed down below. The part was tagged as G-Block 6 and sample 6-2 yielded accurate results. | ||
| − | [[File: | + | [[File:AUC_TURKEY_6_colony.png]] [[File:6004-2.png]] |
[[File:6004-3.png]] [[File:6004-4.png]] | [[File:6004-3.png]] [[File:6004-4.png]] | ||
Revision as of 20:44, 20 September 2015
T7+ ibPB RNA Thermometer + pelB + MCS + 6xHis
The part is to act as a backbone for the T7+ibPB RNA Thermometer+pelB+6xHis system. The MCS(Multiple Cloning Site) as the ligation region for the parts to be integrated into the system. The part that is added to the backbone will be translated in the presence of higher temperatures. The pelB signal peptide is present for the produced protein to be carried out of the cell. The 6xHis is for protein analysis.
Cloning
The composite system was ligated to the cloning vector psB1C3, to be transformated and then verified with Colony PCR. The verified ligation was then moved on to the digestion step to be cut with the EcoR1 and Pst1 restriction enzymes. The gel run of the cut parts were portrayed down below. The part was tagged as G-Block 6 and sample 6-2 yielded accurate results.
Functional Assay
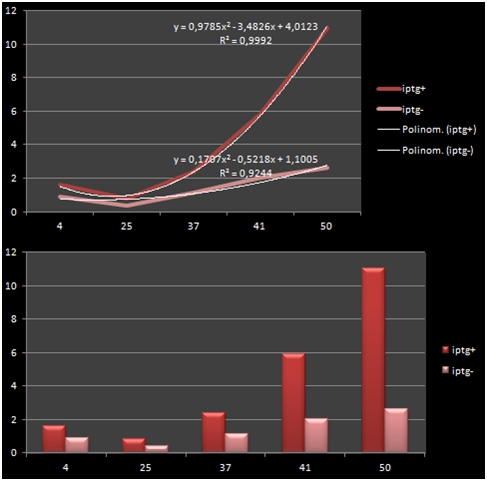
The bacteria that were transformated with G-Block 7 were cultured in 5 ml and incubated for 13 hours. After this incubation, the cultures were incubated at respective temperatures for 4 hours and were given 50uM of iPTG twice with 4 hours periouds. There were a total 5 different temperatures that the cultures were incubated in: 4, 25, 37, 42 and 50 C. The proteins in the cultures were then isolated and the protein concentrations were measured. Data on RFP concentration were acquired at 584 nm of emission and 607 nm of excitation. The acquired values were divided to the total amount of protein to acquire the following ratio.
| Medium Temperature | iPTG Presence +/- | Total Amount of Protein | RFP Fluorometric Measurement | RFP/Total |
|---|---|---|---|---|
| 4 | + | 14.09 | 22.32 | 1.584 |
| 4 | - | 11.349 | 10.36 | 0.912 |
| 25 | + | 17.87 | 14.35 | 0.802 |
| 25 | - | 11.054 | 4,313 | 0,390 |
| 37 | + | 16.808 | 40.2 | 2.391 |
| 37 | - | 15.216 | 17.36 | 1.140 |
| 41 | + | 7.637 | 44.83 | 5.870 |
| 41 | - | 5.557 | 11.13 | 2.002 |
| 50 | + | 5.463 | 60.06 | 10.993 |
| 50 | - | 7.997 | 20.95 | 2.619 |
The graph shows the variance in density concentrations of RFP, caused as a result of the functioning of the iPTG Inducible Promoters in different temperatures. The functioning of the promoters increased cumulatively with increased temperatures.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 2
Illegal BamHI site found at 252
Illegal XhoI site found at 277 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 219
- 1000COMPATIBLE WITH RFC[1000]