Difference between revisions of "Part:BBa K1694034"
SherryNCTU (Talk | contribs) |
SherryNCTU (Talk | contribs) |
||
| Line 8: | Line 8: | ||
[[File:PROCGFPPCR.png|200px|thumb|left|'''Fig.2'''The PCR result of the Pcons+B0034+Lpp-OmpA-N+scFv(anti-EGFR)+B0030+GFP+J61048. The DNA sequence length is around 2000~2200 bp, so the PCR products should appear at 2200~2400 bp.]] | [[File:PROCGFPPCR.png|200px|thumb|left|'''Fig.2'''The PCR result of the Pcons+B0034+Lpp-OmpA-N+scFv(anti-EGFR)+B0030+GFP+J61048. The DNA sequence length is around 2000~2200 bp, so the PCR products should appear at 2200~2400 bp.]] | ||
After assemble the DNA sequences from the basic parts, we recombined each Pcons+B0034+Lpp-OmpA-N+scFv(anti-EGFR)+B0030+GFP+J61048 gene to PSB1C3 backbones and conducted a PCR experiment to check the size of each of the parts. The DNA sequence length of the these parts are around 1900~2100 bp. In this PCR experiment, the PCR products size should be near at 2100~2300 bp. The Fig.2 showed the correct size of this part, and proved that we successful ligated the sequence onto an ideal backbone. | After assemble the DNA sequences from the basic parts, we recombined each Pcons+B0034+Lpp-OmpA-N+scFv(anti-EGFR)+B0030+GFP+J61048 gene to PSB1C3 backbones and conducted a PCR experiment to check the size of each of the parts. The DNA sequence length of the these parts are around 1900~2100 bp. In this PCR experiment, the PCR products size should be near at 2100~2300 bp. The Fig.2 showed the correct size of this part, and proved that we successful ligated the sequence onto an ideal backbone. | ||
| + | |||
| + | <p style="font-size:120%">'''2. Transformation of single plasmid'''</p> | ||
| + | |||
| + | |||
| + | To prove that our scFv can actually bind on to the antigen on cancer cells, we connected each scFv with a different fluorescence protein. Therefore we could use fluorescence microscope to clearly observe if the ''E. coli'' has produced scFv proteins. Currently, we built three different scFv connected with their respectively fluorescence protein. When applied on cell staining, we can identify the antigen distribution on cancer cells by observing the fluorescence. Furthermore, if we use the three scFv simultaneously, we can also detect multiple markers. | ||
| + | |||
| + | <br> | ||
| + | ''' (1) Parts:''' | ||
| + | [[File:TR.png|600px|thumb|center|'''Fig.13''' Transformation of single plasmid]] | ||
| + | [[File:EGFRRFP.png|900px|thumb|center|'''Fig.14''' Pcons+RBS+Lpp-OmpA-N+Anti-EGFR+RBS+RFP+Ter]] | ||
| + | <br> | ||
| + | [[File:EGFRGFP.png|900px|thumb|center|'''Fig.15''' Pcons+RBS+Lpp-OmpA-N+Anti-EGFR+RBS+GFP+Ter]] | ||
| + | <br> | ||
| + | |||
| + | ''' (2) Cell staining experiment:''' | ||
| + | After creating the part of scFv and transforming them into our ''E. coli'', we were going to prove that our detectors have successfully displayed scFv of anti-EGFR. To prove this, we have decided to undergo the cell staining experiment by using our ''E. coli'' to detect the EGFR in the SKOV-3 cancer cell lines. SKOV-3 is a kind of epithelial cell that expressed markers such as EGFR. | ||
| + | <br> | ||
| + | <br> | ||
| + | ''' (3) Staining results:''' | ||
| + | |||
| + | <br> | ||
| + | <div style="display: block; height: 250pt;"> | ||
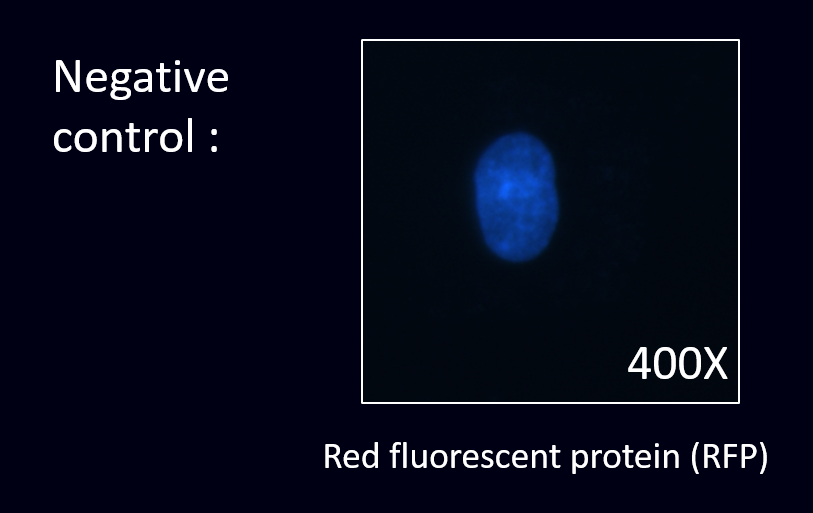
| + | [[File:Co3.png|400px|thumb|left|'''Fig.16''' As results,there is no red fluorescent ''E. coli'' stick on the cell’s surface as there is no specific scFv displayed around the ''E. coli''.]] | ||
| + | [[File:EGFPRFPCELL1.png|400px|thumb|left|'''Fig.17''' There are green fluorescent anti-EGFR ''E. coli'' stick on the cell’s surface as the anti-EGFR probes on ''E. coli'' successfully detect and bind with EGFR.]] | ||
| + | </div> | ||
| + | |||
| + | <div style="display: block; height: 250pt;"> | ||
| + | |||
| + | [[File:Ng.png|400px|thumb|left|'''Fig.18''' As results,there is no green fluorescent ''E. coli'' stick on the cell’s surface as there is no specific scFv displayed around the ''E. coli''.]] | ||
| + | [[File:EGFPGFPCELL1.png|400px|thumb|left|'''Fig.19''' There are green fluorescent anti-EGFR ''E. coli'' stick on the cell’s surface as the anti-EGFR probes on ''E. coli'' successfully detect and bind with EGFR.]] | ||
| + | </div> | ||
| + | <div style="display: block; height: 300pt;"> | ||
| + | [[File:Ng.png|400px|thumb|left|'''Fig.20''' As results,there is no green fluorescent ''E. coli'' stick on the cell’s surface as there is no specific scFv displayed around the ''E. coli''.]] | ||
| + | [[File:EGFPAMILCPCELL.png|400px|thumb|left|'''Fig.21''' There are blue chromoprotein anti-EGFR ''E. coli'' stick on the cell’s surface as the anti-EGFR probes on ''E. coli'' successfully detect and bind with EGFR.]] | ||
| + | </div> | ||
[[File:PROCGFP.png|600px|thumb|center|'''Fig.3''' Pcons+B0034+Lpp-OmpA-N+scFv(anti-EGFR)+B0030+GFP+J61048]] | [[File:PROCGFP.png|600px|thumb|center|'''Fig.3''' Pcons+B0034+Lpp-OmpA-N+scFv(anti-EGFR)+B0030+GFP+J61048]] | ||
Revision as of 19:52, 19 September 2015
Pcons+B0034+Lpp-OmpA-N+scFv(Anti-EGFR)+B0030+GFP+J61048
Introduction:
To prove that our scFv can actually bind on to the antigen on cancer cells, we connected each scFv with a different fluorescence protein. Therefore we could use fluorescence microscope to clearly observe if the E. coli has produced scFv proteins. Currently,we built three different scFv connected with their respectively fluorescence protein, which are Anti-VEGF+GFP, Anti-EGFR+RFP, Anti-Her2+BFP. When applied on cell staining, we can identify the antigen distribution on cancer cells by observing the fluorescence. Furthermore, if we use the three scFv simultaneously, we can also detect multiple markers. Moreover, we built combinations of each scFv connected with GFP.
Experiment
After assemble the DNA sequences from the basic parts, we recombined each Pcons+B0034+Lpp-OmpA-N+scFv(anti-EGFR)+B0030+GFP+J61048 gene to PSB1C3 backbones and conducted a PCR experiment to check the size of each of the parts. The DNA sequence length of the these parts are around 1900~2100 bp. In this PCR experiment, the PCR products size should be near at 2100~2300 bp. The Fig.2 showed the correct size of this part, and proved that we successful ligated the sequence onto an ideal backbone.
2. Transformation of single plasmid
To prove that our scFv can actually bind on to the antigen on cancer cells, we connected each scFv with a different fluorescence protein. Therefore we could use fluorescence microscope to clearly observe if the E. coli has produced scFv proteins. Currently, we built three different scFv connected with their respectively fluorescence protein. When applied on cell staining, we can identify the antigen distribution on cancer cells by observing the fluorescence. Furthermore, if we use the three scFv simultaneously, we can also detect multiple markers.
(1) Parts:
(2) Cell staining experiment:
After creating the part of scFv and transforming them into our E. coli, we were going to prove that our detectors have successfully displayed scFv of anti-EGFR. To prove this, we have decided to undergo the cell staining experiment by using our E. coli to detect the EGFR in the SKOV-3 cancer cell lines. SKOV-3 is a kind of epithelial cell that expressed markers such as EGFR.
(3) Staining results:
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 451
Illegal NgoMIV site found at 2001 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1917