Difference between revisions of "Part:BBa K1679018"
m (→Usage and Biology) |
m |
||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1679018 short</partinfo> | <partinfo>BBa_K1679018 short</partinfo> | ||
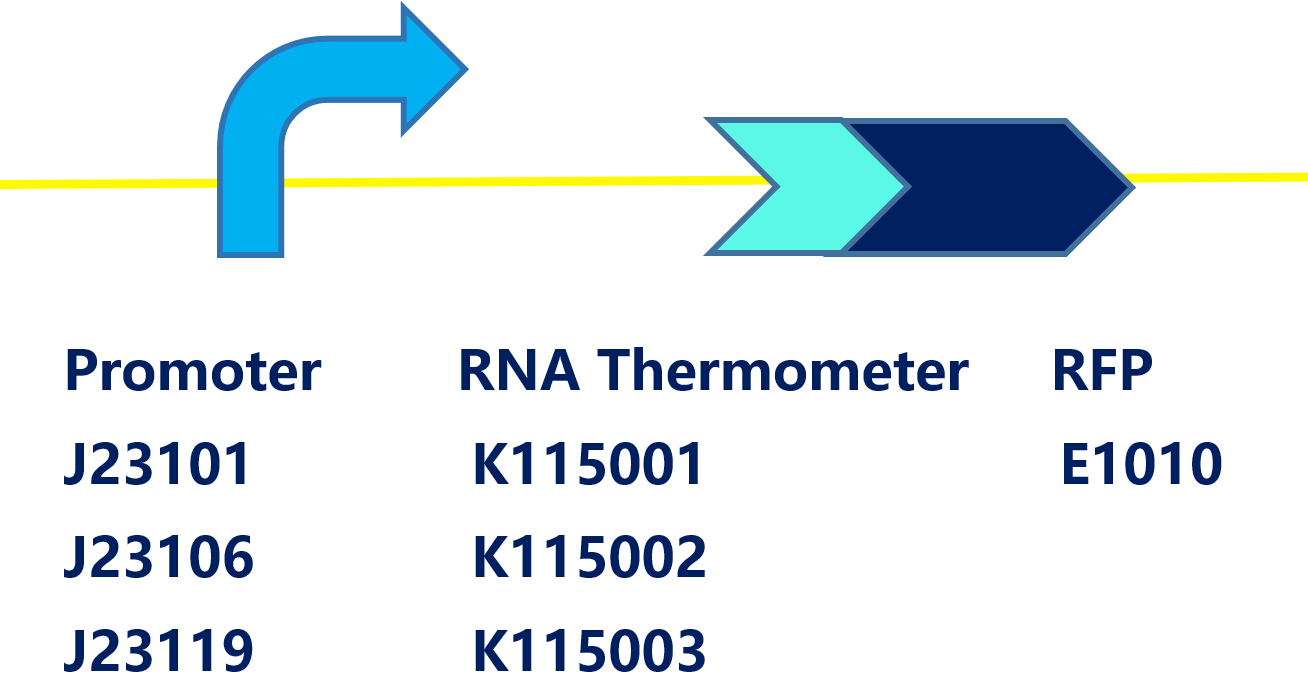
| − | This is a device containing a promoter ( | + | This is a device containing a promoter (BBa_J23101),a RNA thermometer (BBa_K115001) and a RFP coding sequence (BBa_E0010) on the pSB1C3, which can be easily transformed into E.coli. |
| − | RNA Thermometers(RNAT) are temperature-sensing RNA sequences in 5’UTR of their mRNAs. At low temperature, RNAT folds into structure, blocking access of ribosome; | + | RNA Thermometers(RNAT) are temperature-sensing RNA sequences in 5’UTR of their mRNAs. At low temperature, RNAT folds into structure, blocking access of ribosome; at high temperature, RNAT switch from off to open conformation, increasing the efficiency of translation initiation. |
When the temperature rises above 37°C, RFP would be expressed in theory. | When the temperature rises above 37°C, RFP would be expressed in theory. | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | [[File:RNAt.jpg|450px|thumb]] | + | [[File:RNAt.jpg|450px|thumb|centre]] |
| − | [[File:3plateRNAT.png|450px|thumb | + | [[File:3plateRNAT.png|450px|thumb|left| For one temperature, we spotted one strain on 3 different plates, each plate contains 9 different strains. After 48 hours, as we can see in the photo: FourU worked best under all these promoters and show great switch activity when be used cooperatively with J23101 and J23109. Our K1679018 didn't show significant difference between 37℃ and 42℃ conditions.]] |
| − | + | [[File:119-003.jpg|px1500|thumb|centre|These are our K1679018 which were cultured at 28℃, 35℃, 37℃, 40℃, and 42℃ from right to left. It didn't show significant difference between these temperature in liquid LB medium.]] | |
| + | TUDelft-2008 has modified 3 kinds of RNAT: ROSE(BBa_K115001), FourU(BBa_ K115002), PrfA(BBa_K115003). We constructed each RNAT under the control of 3 different constitutive promoters:BBa_J23101, BBa_J23106, BBa_J23119 and use RFP as a reporter. After construction, the 9 circuits were transformed into DH5α. | ||
| − | + | According to the results of TUDelft-2008, the temperature threshold is 37℃ for FourU and PfrA, 42℃ for ROSE. Thus, we set the culture temperature to 28℃, 37℃ and 42℃ on solid LB medium while 28℃, 35℃, 37℃, 40℃, and 42℃ in liquid LB medium. | |
| − | According to the results of TUDelft-2008, the temperature threshold is 37℃ for FourU and PfrA, 42℃ for ROSE. Thus, we set the culture temperature to 28℃, 37℃ and 42℃ | + | |
| − | + | ||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K1679018 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1679018 SequenceAndFeatures</partinfo> | ||
Revision as of 18:29, 18 September 2015
Promoter+RNA thermometer (PrfA) +mRFP
This is a device containing a promoter (BBa_J23101),a RNA thermometer (BBa_K115001) and a RFP coding sequence (BBa_E0010) on the pSB1C3, which can be easily transformed into E.coli. RNA Thermometers(RNAT) are temperature-sensing RNA sequences in 5’UTR of their mRNAs. At low temperature, RNAT folds into structure, blocking access of ribosome; at high temperature, RNAT switch from off to open conformation, increasing the efficiency of translation initiation. When the temperature rises above 37°C, RFP would be expressed in theory.
Usage and Biology

TUDelft-2008 has modified 3 kinds of RNAT: ROSE(BBa_K115001), FourU(BBa_ K115002), PrfA(BBa_K115003). We constructed each RNAT under the control of 3 different constitutive promoters:BBa_J23101, BBa_J23106, BBa_J23119 and use RFP as a reporter. After construction, the 9 circuits were transformed into DH5α.
According to the results of TUDelft-2008, the temperature threshold is 37℃ for FourU and PfrA, 42℃ for ROSE. Thus, we set the culture temperature to 28℃, 37℃ and 42℃ on solid LB medium while 28℃, 35℃, 37℃, 40℃, and 42℃ in liquid LB medium.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 713
Illegal AgeI site found at 825 - 1000COMPATIBLE WITH RFC[1000]