Difference between revisions of "Part:BBa K1618037"
LedaCoelewij (Talk | contribs) |
|||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
| − | + | <partinfo>BBa_K1467101 short</partinfo> | |
| − | + | ||
| − | + | ||
| + | 35S is a plant specific terminator obtained from the Cauliflower Mosaic Virus. The part’s intended use is to terminate transcription in plant cells. This part was converted to a standard BioBrick by cloning into the Mo-Flipper Plasmid K1467300 made by the NRP-UEA 2014 team. | ||
| + | NRP-UEA 2015 team used this part in our plant constructs. Constructs K1618033-36 contained a 35s promoter, chloroplast transit peptide, one of four acyltransferase, and this 35s terminator. Parts K1618029-32 were the same as the first four, but also contained a GFP tag to confirm the localisation to the chloroplast. | ||
| + | |||
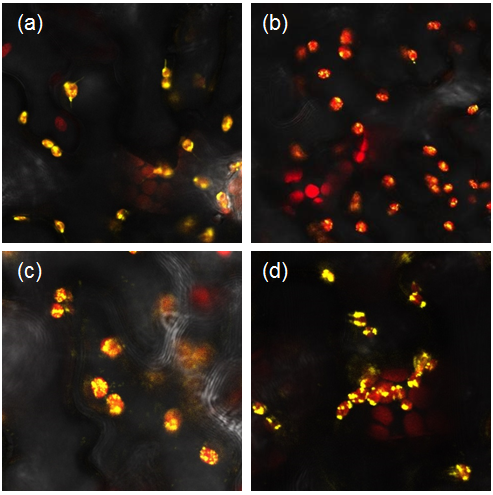
| + | We transformed our constructs into <i>Agrobacterium tumefaciens</i> and then infiltrated it into our plants. We used the YFP tagged constructs to confirm the localisation of parts to the chloroplast (Figure 1) and the untagged constructs to analyse the starch content of our leaves. | ||
| + | |||
| + | |||
| + | [[File:Confocal_Image_029-032.png]] | ||
| + | |||
| + | <i> Figure 1: Constructs BBa_K1618029‐032 contain a yellow fluorescent protein, as well as a chloroplast transit peptide. These are confocal microscopy images of the constructs infiltrated into Nicotiana benthamiana, in which the red structures are the chlorophyll within the chloroplast, and the yellow is the fluorescent fusion protein expressed from constructs (a) BBa_K1618029, (b) BBa_K1618031, (c) BBa_K1618032, and (d) BBa_K1618030. </i> | ||
| + | |||
| + | |||
| + | The above results not only indicate a successful localisation, but also suggest that the 35s terminator that we used is GoldenGate compatible and functioning. | ||
| + | |||
| + | |||
| + | |||
[[Image:Terminator.png]] | [[Image:Terminator.png]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
'''Secondary Structure''' | '''Secondary Structure''' | ||
Revision as of 14:55, 17 September 2015
GoldenGate compatible 35s Promoter
35S is a plant specific terminator obtained from the Cauliflower Mosaic Virus. The part’s intended use is to terminate transcription in plant cells. This part was converted to a standard BioBrick by cloning into the Mo-Flipper Plasmid K1467300 made by the NRP-UEA 2014 team.
NRP-UEA 2015 team used this part in our plant constructs. Constructs K1618033-36 contained a 35s promoter, chloroplast transit peptide, one of four acyltransferase, and this 35s terminator. Parts K1618029-32 were the same as the first four, but also contained a GFP tag to confirm the localisation to the chloroplast.
We transformed our constructs into Agrobacterium tumefaciens and then infiltrated it into our plants. We used the YFP tagged constructs to confirm the localisation of parts to the chloroplast (Figure 1) and the untagged constructs to analyse the starch content of our leaves.
Figure 1: Constructs BBa_K1618029‐032 contain a yellow fluorescent protein, as well as a chloroplast transit peptide. These are confocal microscopy images of the constructs infiltrated into Nicotiana benthamiana, in which the red structures are the chlorophyll within the chloroplast, and the yellow is the fluorescent fusion protein expressed from constructs (a) BBa_K1618029, (b) BBa_K1618031, (c) BBa_K1618032, and (d) BBa_K1618030.
The above results not only indicate a successful localisation, but also suggest that the 35s terminator that we used is GoldenGate compatible and functioning.
Secondary Structure
Measurement
- [http://openwetware.org/wiki/Cconboy:Terminator_Characterization/Results How these parts were measured]