Difference between revisions of "Part:BBa K1467204"
LedaCoelewij (Talk | contribs) (Further Characterization NRP-UEA) |
|||
| Line 16: | Line 16: | ||
| + | |||
| + | == NRP-UEA 2015 == | ||
| + | |||
| + | |||
| + | This year at NRP-UEA, we aimed to develop a prebiotic, which involved the acylation/butyrlation of starch. We aimed to achieve this in plants by using various acyltransferases. We used GFP to tag our constructs in order to confirm the localisation of our parts to the chloroplast, where starch is produced. | ||
| + | |||
| + | This GFP part was included in parts [https://parts.igem.org/Part:BBa_K1618029 BBa_K1618029], [https://parts.igem.org/Part:BBa_K1618030 BBa_K1618030], [https://parts.igem.org/Part:BBa_K1618031 BBa_K1618031], and [https://parts.igem.org/Part:BBa_K1618032 BBa_K1618032] due to its GoldenGate and BioBrick compatibility. | ||
Latest revision as of 13:51, 17 September 2015
GoldenGate compatible Green Flourescent Protein (GFP)
A variation of the part BBa_E0040 that has been made compatible for Golden Gate by removal of internal BsaI and BpiI recognition sequences.
This part, originally isolated from Aequorea victoria, fluoresces under UV light when the gene is expressed. GFP absorbs light at a wavelength of 395nm and emits light at 509nm- producing green florescence. As the florescence is easily detectable under UV light, expression is easily tracked making the protein very useful as a reporter.
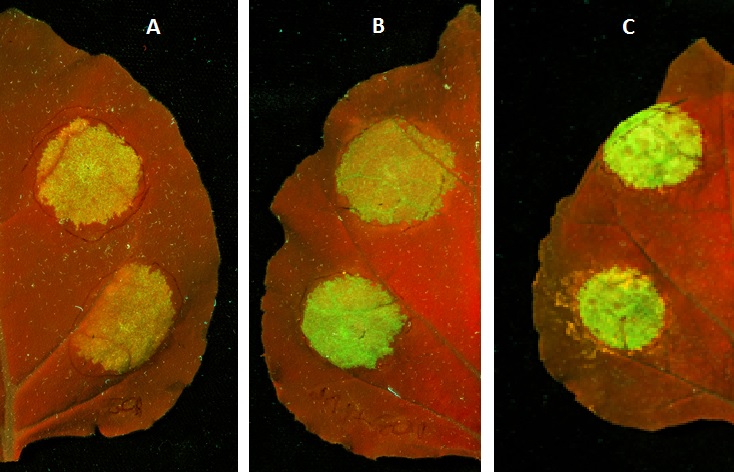
The part was used by iGEM14_NRP-UEA-Norwich as a reporter to test several different promoters with the aim of determining which would be the most useful in further stages of the project. Golden Gate cloning was used to hook GFP up to the promoters 35s (BBa_K1467101), BS3 (BBa_K1467102), PDF1.2 (BBa_K1467103) and PR1 to produce transcriptional units that expressed GFP in Nicotiana Benthamiana following infiltration of agrobacterium carrying the relevant plasmid.
Figure 1: Nicotiana Benthamiana leaves under UV light following infiltration of agrobacterium carrying the constructs (A) 35s_GFP_ocs, (B) BS3_GFP_ocs, (C) PDF1.2_GFP_ocs. In each case GFP protein has be transcribed and is shown as florescence under UV light following activation of the promoter (35s is constitutive, BS3 has been induced by the TALE AvrBS3 and PDF1.2 has been induced by Methyl Jasmonate).
NRP-UEA 2015
This year at NRP-UEA, we aimed to develop a prebiotic, which involved the acylation/butyrlation of starch. We aimed to achieve this in plants by using various acyltransferases. We used GFP to tag our constructs in order to confirm the localisation of our parts to the chloroplast, where starch is produced.
This GFP part was included in parts BBa_K1618029, BBa_K1618030, BBa_K1618031, and BBa_K1618032 due to its GoldenGate and BioBrick compatibility.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]