Difference between revisions of "Part:BBa K1604031"
(→Usage and Biology) |
|||
| Line 6: | Line 6: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | PncB catalyzes one of the rate-limiting step in the NAD synthesis pathway. It has been shown that the overexpression of pncB in <i>E.coli< | + | PncB catalyzes one of the rate-limiting step in the NAD synthesis pathway. It has been shown that the overexpression of pncB in <i>E.coli</i> increases the intracellular level of NAD(1). |
| − | |||
| + | <div style="text-align:center">[[Image:201_3b.jpg]]</div> | ||
| + | <p style="width:600px; margin-left:150px; margin-bottom:60px; | ||
| + | text-align:justify "><b>FIGURE 1. Biochemical pathway of NAD synthesis.</b> PncB gene encodes the transcription of the NARPTase which catalyzes the formation of nicotinate mono-nucleotide from nicotinic acid.</p> | ||
| − | |||
| − | + | <div style="text-align:center">[[Image:201_3b.jpg]]</div> | |
| + | <p style="width:600px; margin-left:150px; margin-bottom:60px; | ||
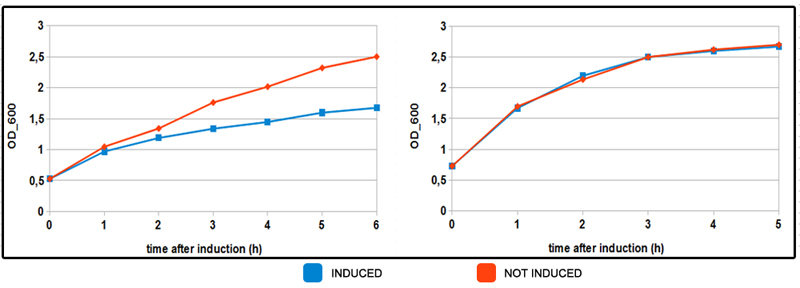
| + | text-align:justify "><b>FIGURE 2. pncB does not affect cells growth rate.</b> Neb10β cells transformed BBa_K1604031 and grown overnight in LB. The day after cells were diluted 1:100 and grown until they reached an optical density (OD600) of 0.5. The cells were splitted in two different aliquotes of 23 mL and induced with 5mM of arabinose. OD was measured every 45 minutes for 5 hours. Negative control were cells transformed with an empty plasmid bearing the arabinose promoter from UniTN Trento 2012 (BBa_K ). All measurements were done for 3 different biological samples and 3 technical measures.</p> | ||
| + | |||
| + | |||
| + | |||
| + | <div style="text-align:center">[[Image:201_3b.jpg]]</div> | ||
| + | <p style="width:600px; margin-left:150px; margin-bottom:60px; | ||
| + | text-align:justify "><b>FIGURE 3. pncB enhances NAD production by ~2.5 fold.</b> Cells expressing pncB (BBa_) and the negative control (BBa_) were grown as described before (figure 2) for a total of 5 hours. After 5 hours the cells the OD was measured again for normalization. 1 mL of cells corresponding to 10<8 were centrifuged and the supernatant was discarded. The NAD and NADH levels were calculated with a colorimetric assay using the Sigma NAD /NADH quantification kit (MAK037) following the instructions of the technical bulletin. Quantification was based on a standard curve with 0, 20, 40, 60, 80 pmole/well of NADH standard. The kit provides the measures of NAD levels indirectly from total levels of NAD + NADH and NADH only. All samples were normalized by removing the blank.</p> | ||
| + | |||
| + | |||
<!-- --> | <!-- --> | ||
Revision as of 10:38, 1 September 2015
araC-pBAD + pncB
pncB encodes for the enzyme NAPRTase (nicotinic acid phosphorbosyl-transferase). It catalyzes the formation of nicotinate mono-nucleotide, a direct precursor of NAD, from NA (nicotinic acid). This device is controlled by an inducible arabinose promoter.
Usage and Biology
PncB catalyzes one of the rate-limiting step in the NAD synthesis pathway. It has been shown that the overexpression of pncB in E.coli increases the intracellular level of NAD(1).
FIGURE 1. Biochemical pathway of NAD synthesis. PncB gene encodes the transcription of the NARPTase which catalyzes the formation of nicotinate mono-nucleotide from nicotinic acid.
FIGURE 2. pncB does not affect cells growth rate. Neb10β cells transformed BBa_K1604031 and grown overnight in LB. The day after cells were diluted 1:100 and grown until they reached an optical density (OD600) of 0.5. The cells were splitted in two different aliquotes of 23 mL and induced with 5mM of arabinose. OD was measured every 45 minutes for 5 hours. Negative control were cells transformed with an empty plasmid bearing the arabinose promoter from UniTN Trento 2012 (BBa_K ). All measurements were done for 3 different biological samples and 3 technical measures.
FIGURE 3. pncB enhances NAD production by ~2.5 fold. Cells expressing pncB (BBa_) and the negative control (BBa_) were grown as described before (figure 2) for a total of 5 hours. After 5 hours the cells the OD was measured again for normalization. 1 mL of cells corresponding to 10<8 were centrifuged and the supernatant was discarded. The NAD and NADH levels were calculated with a colorimetric assay using the Sigma NAD /NADH quantification kit (MAK037) following the instructions of the technical bulletin. Quantification was based on a standard curve with 0, 20, 40, 60, 80 pmole/well of NADH standard. The kit provides the measures of NAD levels indirectly from total levels of NAD + NADH and NADH only. All samples were normalized by removing the blank.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1144
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 979
Illegal AgeI site found at 2317 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 961