Difference between revisions of "Part:BBa K1477014"
Geotwaddle (Talk | contribs) (→Characterization and Application of BBa_K1477014) |
Geotwaddle (Talk | contribs) (→Characterization and Application of BBa_K1477014) |
||
| Line 37: | Line 37: | ||
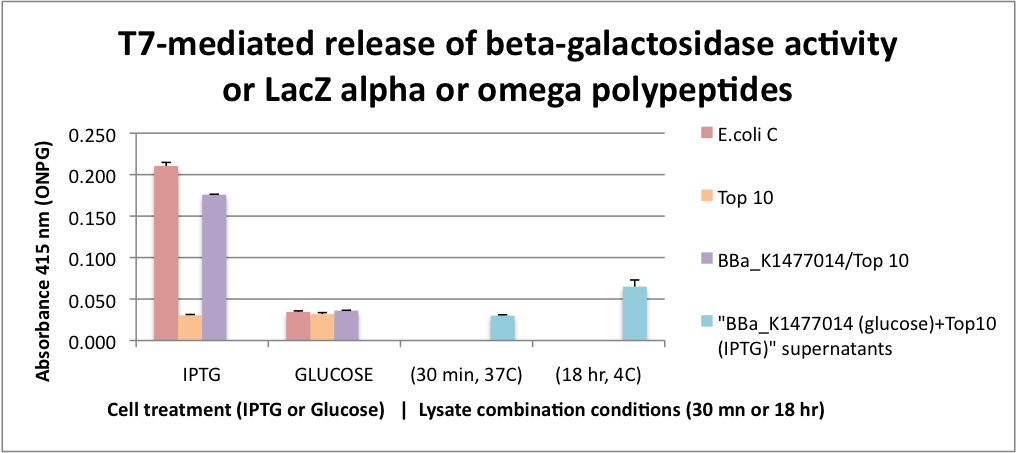
FUNCTIONAL Bgal ALPHA POLYPEPTIDE RELEASED BY CELL LYSIS. This graph represents the results for extracellular alpha complementation of beta galactosidase activity following lytic coliphage liberation of the fragments from transformed cells. To obtain only alpha fragments form BBa_K1477014 we grew the cells on glucose which suppresses the production of the omega polypeptide from the Lac promoter that controls the expression of the delta M15 mutation in the Top 10 host cells. In this assay, we either stimulated beta-gal or omega polypeptide expression with IPTG or suppressed it with glucose. After stimulation or suppression, T7 bacteriophage was added and the cells were incubated for 2 hrs to insure significant cell lysis. Released beta-gal enzyme or beta-gal polypeptide fragments were recovered with SpinX 0.22 micron filter units. Filtrates were combined with ONPG and absorbance was measured at 415 nanometers. | FUNCTIONAL Bgal ALPHA POLYPEPTIDE RELEASED BY CELL LYSIS. This graph represents the results for extracellular alpha complementation of beta galactosidase activity following lytic coliphage liberation of the fragments from transformed cells. To obtain only alpha fragments form BBa_K1477014 we grew the cells on glucose which suppresses the production of the omega polypeptide from the Lac promoter that controls the expression of the delta M15 mutation in the Top 10 host cells. In this assay, we either stimulated beta-gal or omega polypeptide expression with IPTG or suppressed it with glucose. After stimulation or suppression, T7 bacteriophage was added and the cells were incubated for 2 hrs to insure significant cell lysis. Released beta-gal enzyme or beta-gal polypeptide fragments were recovered with SpinX 0.22 micron filter units. Filtrates were combined with ONPG and absorbance was measured at 415 nanometers. | ||
| − | Beta-gal activity was recovered from both E. coli C and | + | Beta-gal activity was recovered from both E. coli C and BBa_K1477014 in ToP10 cells stimulated with IPTG and this activity was surpressed by glucose. In the case of BBa_K1477014 in ToP10 cells, the suppression of beta-gal activity was because of the inhibition of the production of the omega polypeptide. The alpha polypeptide in BBa_K1477014 in ToP10 cells is always produced because it is on a constitutive promoter. |
| − | To test whether alpha complementation could occur extracellularly, a cell filtrate of IPTG Top 10 cells were combined with a cell filtrate from | + | To test whether alpha complementation could occur extracellularly, a cell filtrate of IPTG Top 10 cells were combined with a cell filtrate from BBa_K1477014 in ToP10 cells grown on glucose. β-gal activity unfortunately was not detected in this assay. However we found that when the filtrates were combined and incubated at 4C for 18hrs β-gal activity was detectable suggesting that extracellular complementation had occurred at a slow rate. |
</P> | </P> | ||
Revision as of 01:31, 3 November 2014
Constitutive promoter +RBS+LacZ alpha+Term.
Constitutive promoter (J231202)RBS with the beta-galactosidase alpha fragment peptide coding unit(K1477014).This part is for synthesis and purification of the alpha fragment. It is not designed for blue/white screening.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization and Application of BBa_K1477014
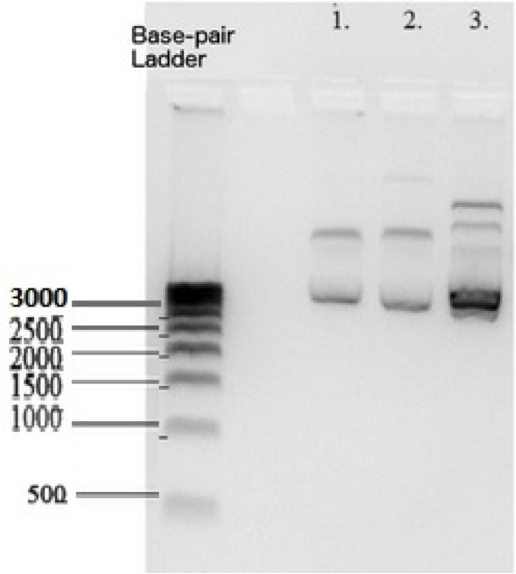
According to the Biorad 500 basepair DNA ladder that we used , K1477014 measures at ~2700 base pairs within pSB1C3 backbone. This matches the sum of the basepairs of individual parts in the device and the backbone. Plasmid DNA isolated from 3 subclones.
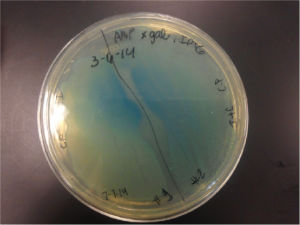
Top10 cells transformed with BBa_K1477014 form blue-green colonies on agar containing X-gal, IPTG, and selective antibiotic confirming the production of a functional β-gal alpha polypeptide and alpha complementation occuring in situ. (2 subclones)
FUNCTIONAL Bgal ALPHA POLYPEPTIDE RELEASED BY CELL LYSIS. This graph represents the results for extracellular alpha complementation of beta galactosidase activity following lytic coliphage liberation of the fragments from transformed cells. To obtain only alpha fragments form BBa_K1477014 we grew the cells on glucose which suppresses the production of the omega polypeptide from the Lac promoter that controls the expression of the delta M15 mutation in the Top 10 host cells. In this assay, we either stimulated beta-gal or omega polypeptide expression with IPTG or suppressed it with glucose. After stimulation or suppression, T7 bacteriophage was added and the cells were incubated for 2 hrs to insure significant cell lysis. Released beta-gal enzyme or beta-gal polypeptide fragments were recovered with SpinX 0.22 micron filter units. Filtrates were combined with ONPG and absorbance was measured at 415 nanometers. Beta-gal activity was recovered from both E. coli C and BBa_K1477014 in ToP10 cells stimulated with IPTG and this activity was surpressed by glucose. In the case of BBa_K1477014 in ToP10 cells, the suppression of beta-gal activity was because of the inhibition of the production of the omega polypeptide. The alpha polypeptide in BBa_K1477014 in ToP10 cells is always produced because it is on a constitutive promoter. To test whether alpha complementation could occur extracellularly, a cell filtrate of IPTG Top 10 cells were combined with a cell filtrate from BBa_K1477014 in ToP10 cells grown on glucose. β-gal activity unfortunately was not detected in this assay. However we found that when the filtrates were combined and incubated at 4C for 18hrs β-gal activity was detectable suggesting that extracellular complementation had occurred at a slow rate.