Difference between revisions of "Part:BBa K1499252"
(→Usage and Biology) |
|||
| Line 8: | Line 8: | ||
This construct is used to validate our amberless expression system, an orthogonal translation system that uses an amber-suppressing tRNA to incorporate leucine into proteins at sites coded for by the amber stop codon (UAG). This system works to mitigate the effects of horizontal gene transfer by ensuring proper expression only in cells a) carrying the tRNA and b) lacking the release factor responsible for ending translation at a UAG (RF1). We have combined our GFP construct (which differs from E0040 at only two sites, changing normal leucine codons to UAGs) with a well-categorized constitutive promoter (J23104), RBS (B0034), and terminator (B0010). Added to the end of these four parts is the supP tRNA, from Stanford-Brown-Spelman's 2014 Amberless toolkit, or [https://parts.igem.org/Part:BBa_K1499251 BBa_K1499251]. | This construct is used to validate our amberless expression system, an orthogonal translation system that uses an amber-suppressing tRNA to incorporate leucine into proteins at sites coded for by the amber stop codon (UAG). This system works to mitigate the effects of horizontal gene transfer by ensuring proper expression only in cells a) carrying the tRNA and b) lacking the release factor responsible for ending translation at a UAG (RF1). We have combined our GFP construct (which differs from E0040 at only two sites, changing normal leucine codons to UAGs) with a well-categorized constitutive promoter (J23104), RBS (B0034), and terminator (B0010). Added to the end of these four parts is the supP tRNA, from Stanford-Brown-Spelman's 2014 Amberless toolkit, or [https://parts.igem.org/Part:BBa_K1499251 BBa_K1499251]. | ||
| − | + | [[Image:SBSiGEM_HellCell1.png|600px|thumb|center|<b>Figure 1.</b> Our approach to the Amberless Codon Security. By recoding the UAG stop codon to translate into an amino acid, only cells that have a tRNA with the anticodon AUC will produce the complete protein. In our experiment, we used a tRNA that charges with leucine to translate the UAG codon.]] | |
| − | [[Image:SBS_AmberlessOverview2.png|700px|thumb|center|<b>Figure | + | |
| + | The complete generator was used to test the orthogonality of the UAG->Leucine coding (Figure 2). | ||
| + | |||
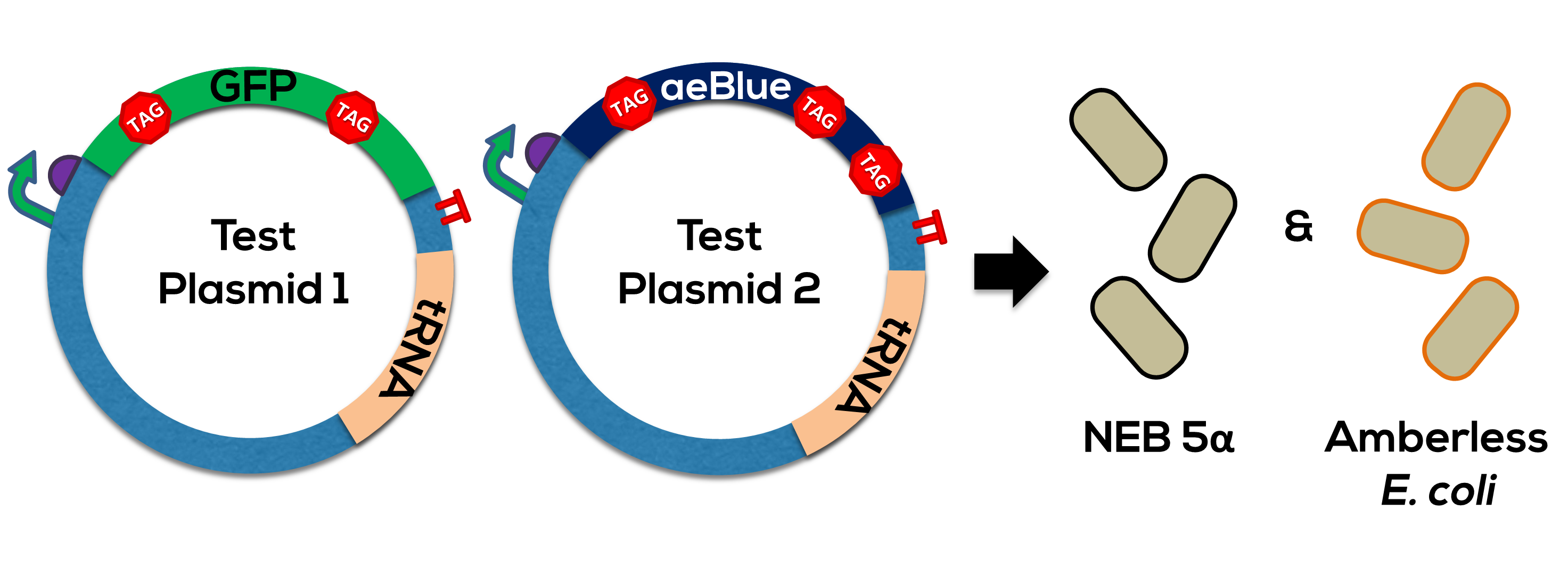
| + | [[Image:SBS_AmberlessOverview2.png|700px|thumb|center|<b>Figure 2.</b> We transformed DH5-alpha and amberless cells with test plasmids, in this case the GFP reporter gene with stop codons and the supP tRNA, in order to establish a proof-of-concept for orthogonality using Codon Security.]] | ||
We have observed that this generator works well in amberless ''E. coli'', but not DH5-alpha. Because of the toxicity of the tRNA, the part will be difficult to transform into any strain of bacterium that contains genes with the amber stop codon. | We have observed that this generator works well in amberless ''E. coli'', but not DH5-alpha. Because of the toxicity of the tRNA, the part will be difficult to transform into any strain of bacterium that contains genes with the amber stop codon. | ||
| Line 18: | Line 21: | ||
<partinfo>BBa_K1499252 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1499252 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | ===Verification=== | ||
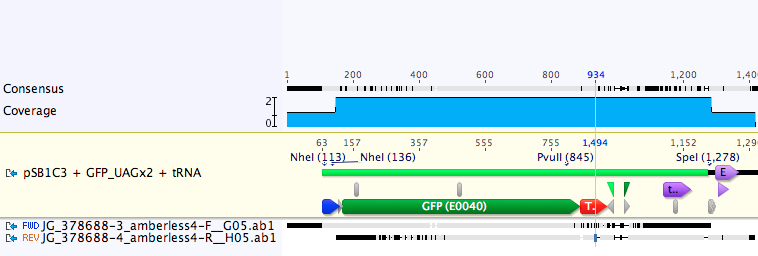
| + | The part was sequence verified in the pSB1C3 backbone before submission to the registry. Two reads, forward and reverse, were obtained using VF2 and VR (Figure 3). | ||
| + | |||
| + | [[Image:UAG_Seq.png|700px|thumb|center|<b>Figure 3.</b> GFP with UAG stop codons and supP tRNA match expected sequence. The GFP goes from bp 167 to 888.The tRNA goes from bp 1141 to 1225.]] | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Revision as of 22:07, 2 November 2014
GFP with 2 stop codons generator + supP tRNA
This composite part combines an ambered GFP generator (K1499250, 2 amber stop codons) with the amber-suppressing tRNA supP (mutated LeuX, K1499251).
Usage and Biology
This construct is used to validate our amberless expression system, an orthogonal translation system that uses an amber-suppressing tRNA to incorporate leucine into proteins at sites coded for by the amber stop codon (UAG). This system works to mitigate the effects of horizontal gene transfer by ensuring proper expression only in cells a) carrying the tRNA and b) lacking the release factor responsible for ending translation at a UAG (RF1). We have combined our GFP construct (which differs from E0040 at only two sites, changing normal leucine codons to UAGs) with a well-categorized constitutive promoter (J23104), RBS (B0034), and terminator (B0010). Added to the end of these four parts is the supP tRNA, from Stanford-Brown-Spelman's 2014 Amberless toolkit, or BBa_K1499251.
The complete generator was used to test the orthogonality of the UAG->Leucine coding (Figure 2).
We have observed that this generator works well in amberless E. coli, but not DH5-alpha. Because of the toxicity of the tRNA, the part will be difficult to transform into any strain of bacterium that contains genes with the amber stop codon.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 705
Verification
The part was sequence verified in the pSB1C3 backbone before submission to the registry. Two reads, forward and reverse, were obtained using VF2 and VR (Figure 3).