Difference between revisions of "Part:BBa K1471009"
| Line 41: | Line 41: | ||
Poly(A) tails enhance the stability and translation of most eukaryotic messenger RNAs, but difficulties in globally measuring poly(A)-tail lengths have impeded greater understanding of poly(A)-tail function. Here we describe poly(A)-tail length profiling by sequencing (PAL-seq) and apply it to measure tail lengths of millions of individual RNAs isolated from yeasts, cell lines, Arabidopsis thaliana leaves, mouse liver, and zebrafish and frog embryos. Poly(A)-tail lengths were conserved between orthologous mRNAs, with mRNAs encoding ribosomal proteins and other ‘housekeeping’ proteins tending to have shorter tails. | Poly(A) tails enhance the stability and translation of most eukaryotic messenger RNAs, but difficulties in globally measuring poly(A)-tail lengths have impeded greater understanding of poly(A)-tail function. Here we describe poly(A)-tail length profiling by sequencing (PAL-seq) and apply it to measure tail lengths of millions of individual RNAs isolated from yeasts, cell lines, Arabidopsis thaliana leaves, mouse liver, and zebrafish and frog embryos. Poly(A)-tail lengths were conserved between orthologous mRNAs, with mRNAs encoding ribosomal proteins and other ‘housekeeping’ proteins tending to have shorter tails. | ||
The poly(A) tail also protects the mRNA from nucleases, in the course of the life of a mRNA, the poly(A) tail is systematically shortened by poly(A)-specific nucleases. Once the poly(A) tail is too short to allow binding of PAB, the circular structure shown above unravels, and both the cap and mRNA 3’ end become “exposed”. | The poly(A) tail also protects the mRNA from nucleases, in the course of the life of a mRNA, the poly(A) tail is systematically shortened by poly(A)-specific nucleases. Once the poly(A) tail is too short to allow binding of PAB, the circular structure shown above unravels, and both the cap and mRNA 3’ end become “exposed”. | ||
| + | |||
| + | |||
| + | '''Reference''' | ||
| + | Taiz,L. and E.Zeiger.(2006) Fisiologia Vegetal.Universitat Jaume. 3rd Edition.Pages 7-27 | ||
| + | |||
| + | Rangan, et al. (2008). Analysis of Context Sequence Surrounding Translation Initiation Site from Complete Genome of | ||
| + | |||
| + | Model Plants. New York University. [Online] Retrieved october 14th 2014 from: http://www.nyu.edu/projects/vogel/Reprints/Rangan_MolBt08.pdf Nakagawa, et al. (2007). Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Oxford University Press. [Online] Retrieved october 14th 2014 from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2241899/ Liu Q, Xue Q. (2005). | ||
| + | |||
| + | Comparative studies on sequence characteristics around translation initiation codon in four eukaryotes. Zhejiang University. [Online] Retrieved october 14th 2014 from: http://www.ias.ac.in/jgenet/Vol84No3/317.pdf Kozak, M. (1989). Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. | ||
| + | |||
| + | American Society for Microbiology (ASM). [Online] retrieved october 14th 2014 from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC363665/ | ||
| + | |||
| + | Classical Nuclear Localization Signals: Definition, Function, and Interaction with Importin α*. (n.d.). Classical Nuclear Localization Signals: Definition, Function, and Interaction with Importin α. Retrieved October 11, 2014, from http://www.jbc.org/content/282/8/5101.full | ||
| + | |||
| + | Nuclear localization sequence. (2014, November 10). Wikipedia. Retrieved October 11, 2014, from http://en.wikipedia.org/wiki/Nuclear_localization_sequence Result Filters. (n.d.). | ||
| + | |||
| + | National Center for Biotechnology Information. Retrieved October 11, 2014, from http://www.ncbi.nlm.nih.gov/pubmed/8241603 Poly(A)-tail profiling reveals an embryonic switch in translational | ||
| + | |||
| + | control. (n.d.). Nature journal. Retrieved October 11, 2014, from http://www.nature.com/nature/journal/v508/n7494/full/nature13007.html | ||
| + | |||
| + | The Poly(A) Tail. (n.d.). The RNA Underworld. Retrieved October 12, 2014, from http://aghunt.wordpress.com/2008/06/05/the-polya-tail/ | ||
| + | poly-A tail. (n.d.). Nature.com. Retrieved October 12, 2014, from http://www.nature.com/scitable/definition/poly-a-tail-276 | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 16:23, 1 November 2014
RBS - PhyB - AGS - VP16 - NLS - PolyA.
RBS
The initiation of protein biosynthesis is a major determinant of the efficiency of gene expression at the translational level. It is known that the nucleotide sequences around the AUG translation initiation codon act as an important signal to trigger the initiation of the translation event. (Kozak, 1987) Understanding regulatory mechanisms of protein synthesis in eukaryotes is essential for the accurate annotation of genome sequences. Kozak reported that the nucleotide sequence GCCGCC(A/G)CCAUGG (AUG is the initiation codon) was frequently observed in vertebrate genes and that this 'consensus' sequence enhanced translation initiation. However, later studies using invertebrate, fungal and plant genes reported different 'consensus' sequences. (Nakagawa, 2007)
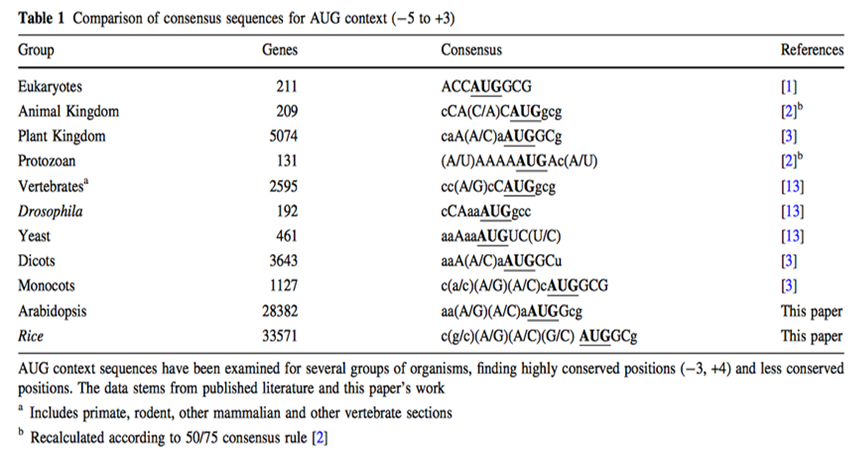
Although for any protein analysis it is crucial to know exactly which region of the mRNA is coding for protein, prediction of the translation initial site is still an unsolved problem. In eukaryotes, the scanning model postulates that the ribosome attaches first to the 5' end of the mRNA and scans along the 5'-3' direction until it encounters the first AUG. While translation initiation from the first AUG holds true in many cases, there are also a considerable number of exceptions. In these exceptions the main determining factor in AUG choice is the context of the respective codon. (Rangan, 2008) Two decades ago, a consensus sequence for the context of the AUG codon in higher plants was proposed on basis of very limited number of sequences. Joshi and colleagues got the generally assumption that the consensus sequence found (aaaaacaA(A/C)aAUGG) is valid for all plant clades, but Rangan found out that a considerable degree of variation between plants and major between the major eukaryotic groups along with some conserved features. However, the large variability and the periodicity suggest that general structural features rather than precise nucleotide sequence may play an important role in transcription initial site. (Rangan, 2008)
PhyB
Phytochrome B is a photoreceptor protein which belongs to a multigenetic family that goes from Phy A to Phy E, every one of this genes is involved in a different function in the plant. This light-receptor protein is stimulated mostly for red light and far red-light in the spectrum at 700-800nm. Phytochrom B has the ability to absorb the red light in order to be able to absorb the far red light, this happens due to a conformation change in the protein which goes back to its initial state once it absorbs the far red light. The previous conformations are known as Pr which is able to absorb the red light and pfr that absorbs the far red light. Phytochrome B is known to be involved in the seeds germination under certain light conditions, in fact mutants with absence of Phy B does not germinate in the same proportion as the wild type, 80% of the seeds tried to be germinated in absence of light did not show any results.

AGS
AGS linker is a small sequence of aminoacids (3 aa). Its name is derived from the aminoacids components of its chain, which are Alanine (A), Glycine (G) and Serine (S). It's widely used for fusing proteins domains in Tandem-like synthetic constructs.
VP16
VP16 is a transcription factor from Herpes simplex virus that activates the transcription of some viral late genes. In synthetic constructions, VP16 activation domain has probed to be effective at activating the expression of some genes when fused to an adecuate DNA-Binding domain. This part may be useful to simplify the designing of synthetic constructs intended to be used in eucaryotic cells.
NLS
The best understood system for the transport of macromolecules between the cytoplasm and the nucleus is nuclear import pathway. In this pathway, a protein containing a basic nuclear localization signal (NLS) is imported by a heterodimeric import receptor. This nuclear localization signal or sequence (NLS) is an amino acid sequence that tags a protein for import into the cell nucleus by nuclear transport.Typically, this signal consists of one or more short sequences of positively charged lysines or arginines exposed on the protein surface. Different nuclear localized proteins may share the same NLS. The active NLS needs to be exposed to the protein surface, the cell has invented mechanisms to expose a hidden or cryptic NLS by protein phosphorylation, dephosphorylation, dissociation of an inhibitory subunit that masks the NLS, processing of a larger precursor, and binding of a hormone to regulate the nuclear import of a protein transcription factor at a certain stage of development, or cell cycle. It is proposed that a hexapeptide with four arginines and lysines (and histidines, in some cases) is a good candidate for a "core NLS," that acidic domains on proteins to be imported may participate in anchoring them to the transporter cytoplasmic or pore complex NLS-receptor, and that NLS have both a cytoplasmic and a nuclear function. The interaction between nuclear proteins and transporter proteins in the pore appears to be largely electrostatic and to be disrupted by the binding of mRNA molecules to the same transporter protein, functioning also for the cytoplasmic export of RNA.
Poly-A
The poly-A tail is a long chain of adenine nucleotides that is added to a messenger RNA (mRNA) molecule during RNA processing to increase the stability of the molecule. Immediately after a gene in a eukaryotic cell is transcribed, the new RNA molecule undergoes several modifications known as RNA processing. These modifications alter both ends of the primary RNA transcript to produce a mature mRNA molecule. The processing of the 3' end adds a poly-A tail to the RNA molecule. First, the 3' end of the transcript is cleaved to free a 3' hydroxyl. Then an enzyme called poly-A polymerase adds a chain of adenine nucleotides to the RNA. This process, called polyadenylation, adds a poly-A tail that is between 100 and 250 residues long. The poly-A tail makes the RNA molecule more stable and prevents its degradation. Additionally, the poly-A tail allows the mature messenger RNA molecule to be exported from the nucleus and translated into a protein by ribosomes in the cytoplasm. Poly(A) tails enhance the stability and translation of most eukaryotic messenger RNAs, but difficulties in globally measuring poly(A)-tail lengths have impeded greater understanding of poly(A)-tail function. Here we describe poly(A)-tail length profiling by sequencing (PAL-seq) and apply it to measure tail lengths of millions of individual RNAs isolated from yeasts, cell lines, Arabidopsis thaliana leaves, mouse liver, and zebrafish and frog embryos. Poly(A)-tail lengths were conserved between orthologous mRNAs, with mRNAs encoding ribosomal proteins and other ‘housekeeping’ proteins tending to have shorter tails. The poly(A) tail also protects the mRNA from nucleases, in the course of the life of a mRNA, the poly(A) tail is systematically shortened by poly(A)-specific nucleases. Once the poly(A) tail is too short to allow binding of PAB, the circular structure shown above unravels, and both the cap and mRNA 3’ end become “exposed”.
Reference
Taiz,L. and E.Zeiger.(2006) Fisiologia Vegetal.Universitat Jaume. 3rd Edition.Pages 7-27
Rangan, et al. (2008). Analysis of Context Sequence Surrounding Translation Initiation Site from Complete Genome of
Model Plants. New York University. [Online] Retrieved october 14th 2014 from: http://www.nyu.edu/projects/vogel/Reprints/Rangan_MolBt08.pdf Nakagawa, et al. (2007). Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Oxford University Press. [Online] Retrieved october 14th 2014 from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2241899/ Liu Q, Xue Q. (2005).
Comparative studies on sequence characteristics around translation initiation codon in four eukaryotes. Zhejiang University. [Online] Retrieved october 14th 2014 from: http://www.ias.ac.in/jgenet/Vol84No3/317.pdf Kozak, M. (1989). Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs.
American Society for Microbiology (ASM). [Online] retrieved october 14th 2014 from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC363665/
Classical Nuclear Localization Signals: Definition, Function, and Interaction with Importin α*. (n.d.). Classical Nuclear Localization Signals: Definition, Function, and Interaction with Importin α. Retrieved October 11, 2014, from http://www.jbc.org/content/282/8/5101.full
Nuclear localization sequence. (2014, November 10). Wikipedia. Retrieved October 11, 2014, from http://en.wikipedia.org/wiki/Nuclear_localization_sequence Result Filters. (n.d.).
National Center for Biotechnology Information. Retrieved October 11, 2014, from http://www.ncbi.nlm.nih.gov/pubmed/8241603 Poly(A)-tail profiling reveals an embryonic switch in translational
control. (n.d.). Nature journal. Retrieved October 11, 2014, from http://www.nature.com/nature/journal/v508/n7494/full/nature13007.html
The Poly(A) Tail. (n.d.). The RNA Underworld. Retrieved October 12, 2014, from http://aghunt.wordpress.com/2008/06/05/the-polya-tail/ poly-A tail. (n.d.). Nature.com. Retrieved October 12, 2014, from http://www.nature.com/scitable/definition/poly-a-tail-276
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 507
Illegal BamHI site found at 589
Illegal XhoI site found at 540
Illegal XhoI site found at 559 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 756
