Difference between revisions of "Part:BBa K1471006"
| Line 33: | Line 33: | ||
Taiz,L. and E.Zeiger.(2006) Fisiologia Vegetal.Universitat Jaume. 3rd Edition.Pages 7-27 | Taiz,L. and E.Zeiger.(2006) Fisiologia Vegetal.Universitat Jaume. 3rd Edition.Pages 7-27 | ||
| + | |||
Rangan, et al. (2008). Analysis of Context Sequence Surrounding Translation Initiation Site from Complete Genome of Model Plants. New York University. [Online] Retrieved october 14th 2014 from: http://www.nyu.edu/projects/vogel/Reprints/Rangan_MolBt08.pdf Nakagawa, et al. (2007). Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Oxford University Press. [Online] Retrieved october 14th 2014 from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2241899/ Liu Q, Xue Q. (2005). Comparative studies on sequence characteristics around translation initiation codon in four eukaryotes. Zhejiang University. [Online] Retrieved october 14th 2014 from: http://www.ias.ac.in/jgenet/Vol84No3/317.pdf Kozak, M. (1989). Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. American Society for Microbiology (ASM). [Online] retrieved october 14th 2014 from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC363665/ | Rangan, et al. (2008). Analysis of Context Sequence Surrounding Translation Initiation Site from Complete Genome of Model Plants. New York University. [Online] Retrieved october 14th 2014 from: http://www.nyu.edu/projects/vogel/Reprints/Rangan_MolBt08.pdf Nakagawa, et al. (2007). Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Oxford University Press. [Online] Retrieved october 14th 2014 from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2241899/ Liu Q, Xue Q. (2005). Comparative studies on sequence characteristics around translation initiation codon in four eukaryotes. Zhejiang University. [Online] Retrieved october 14th 2014 from: http://www.ias.ac.in/jgenet/Vol84No3/317.pdf Kozak, M. (1989). Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. American Society for Microbiology (ASM). [Online] retrieved october 14th 2014 from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC363665/ | ||
Revision as of 16:20, 1 November 2014
RBS with PhyB.
RBS for yeast and a light receptor named phytochrome B.
RBS
The initiation of protein biosynthesis is a major determinant of the efficiency of gene expression at the translational level. It is known that the nucleotide sequences around the AUG translation initiation codon act as an important signal to trigger the initiation of the translation event. (Kozak, 1987) Understanding regulatory mechanisms of protein synthesis in eukaryotes is essential for the accurate annotation of genome sequences. Kozak reported that the nucleotide sequence GCCGCC(A/G)CCAUGG (AUG is the initiation codon) was frequently observed in vertebrate genes and that this 'consensus' sequence enhanced translation initiation. However, later studies using invertebrate, fungal and plant genes reported different 'consensus' sequences. (Nakagawa, 2007)
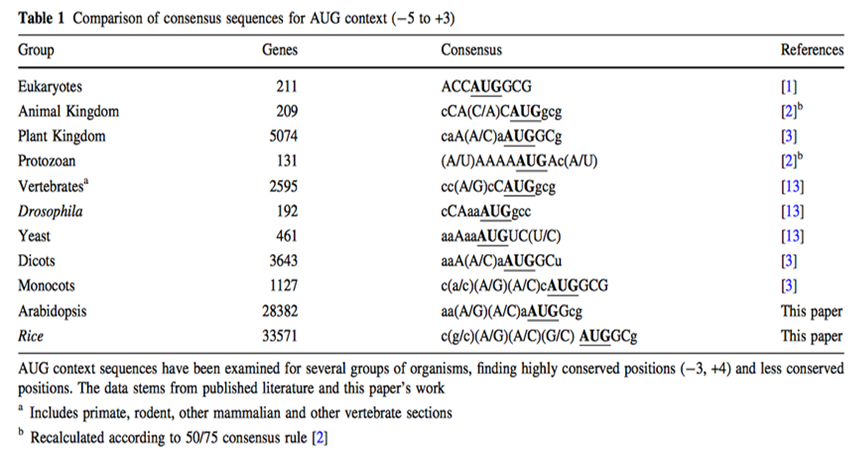
Although for any protein analysis it is crucial to know exactly which region of the mRNA is coding for protein, prediction of the translation initial site is still an unsolved problem. In eukaryotes, the scanning model postulates that the ribosome attaches first to the 5' end of the mRNA and scans along the 5'-3' direction until it encounters the first AUG. While translation initiation from the first AUG holds true in many cases, there are also a considerable number of exceptions. In these exceptions the main determining factor in AUG choice is the context of the respective codon. (Rangan, 2008) Two decades ago, a consensus sequence for the context of the AUG codon in higher plants was proposed on basis of very limited number of sequences. Joshi and colleagues got the generally assumption that the consensus sequence found (aaaaacaA(A/C)aAUGG) is valid for all plant clades, but Rangan found out that a considerable degree of variation between plants and major between the major eukaryotic groups along with some conserved features. However, the large variability and the periodicity suggest that general structural features rather than precise nucleotide sequence may play an important role in transcription initial site. (Rangan, 2008)
PhyB
Phytochrome B is a photoreceptor protein which belongs to a multigenetic family that goes from Phy A to Phy E, every one of this genes is involved in a different function in the plant. This light-receptor protein is stimulated mostly for red light and far red-light in the spectrum at 700-800nm. Phytochrom B has the ability to absorb the red light in order to be able to absorb the far red light, this happens due to a conformation change in the protein which goes back to its initial state once it absorbs the far red light. The previous conformations are known as Pr which is able to absorb the red light and pfr that absorbs the far red light. Phytochrome B is known to be involved in the seeds germination under certain light conditions, in fact mutants with absence of Phy B does not germinate in the same proportion as the wild type, 80% of the seeds tried to be germinated in absence of light did not show any results.
 Figure no. 1
Figure no. 1
In the figure no. 1 its shown the action of the protein under different red light emissions, Where the normal conformation of the protein is change by the red-light emission at 660 nm and then is able to absorb the far red-light and also interact with a specific molecule called phytochrome interacting factor 6 or "pif6" which acts as a signal transmitter from the Phytochrome B to other systems.
In the previous figure no. 2 is shown how the Phy B acts under the constant stimulation of red-light and far red-light, also the action of Phy A under constant light exposure and with low light exposure.
Phytochrome B is known to have a crucial role for plants growing under low exposition to light, also its conformations have a duo regulative action which the pfr conformation. When red-light is absorbed the phytochrome turns into the Pfr conformation which gains the ability to absorb the far red-light and come back to the original conformation Pr.
Phytochrome B mutant lack of chlorophyll molecules, coding genes for proteins involved in the development of chloroplasts and alters the plant capacity for responding to certain vegetal hormones. Also it affects the response ability to red light, the other phytochrome in the plants can´t fulfill the entire function of absorbing red light and white light.
Reference
Taiz,L. and E.Zeiger.(2006) Fisiologia Vegetal.Universitat Jaume. 3rd Edition.Pages 7-27
Rangan, et al. (2008). Analysis of Context Sequence Surrounding Translation Initiation Site from Complete Genome of Model Plants. New York University. [Online] Retrieved october 14th 2014 from: http://www.nyu.edu/projects/vogel/Reprints/Rangan_MolBt08.pdf Nakagawa, et al. (2007). Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Oxford University Press. [Online] Retrieved october 14th 2014 from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2241899/ Liu Q, Xue Q. (2005). Comparative studies on sequence characteristics around translation initiation codon in four eukaryotes. Zhejiang University. [Online] Retrieved october 14th 2014 from: http://www.ias.ac.in/jgenet/Vol84No3/317.pdf Kozak, M. (1989). Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. American Society for Microbiology (ASM). [Online] retrieved october 14th 2014 from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC363665/
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 507
Illegal BamHI site found at 589
Illegal XhoI site found at 540
Illegal XhoI site found at 559 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 756
