Difference between revisions of "Part:BBa K1351019"
| Line 2: | Line 2: | ||
<partinfo>BBa_K1351019 short</partinfo> | <partinfo>BBa_K1351019 short</partinfo> | ||
| − | Artificial constructed complementary RNA which should bind the mRNA of the SdpI immunity protein [https://parts.igem.org/Part:BBa_K1351017 (BBa_K1351017)] in ''Bacillus subtilis'' in order to disturb the translation of immunity. Expected was a loss of immunity in ''B. subtilis'' against its own cannibalism toxin [https://parts.igem.org/Part:BBa_K1351043 | + | Artificial constructed complementary RNA which should bind the mRNA of the SdpI immunity protein [https://parts.igem.org/Part:BBa_K1351017 (BBa_K1351017)] in ''Bacillus subtilis'' in order to disturb the translation of immunity. Expected was a loss of immunity in ''B. subtilis'' against its own cannibalism toxin [https://parts.igem.org/Part:BBa_K1351043 SDP (BBa_K1351043)] over time. |
| Line 21: | Line 21: | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
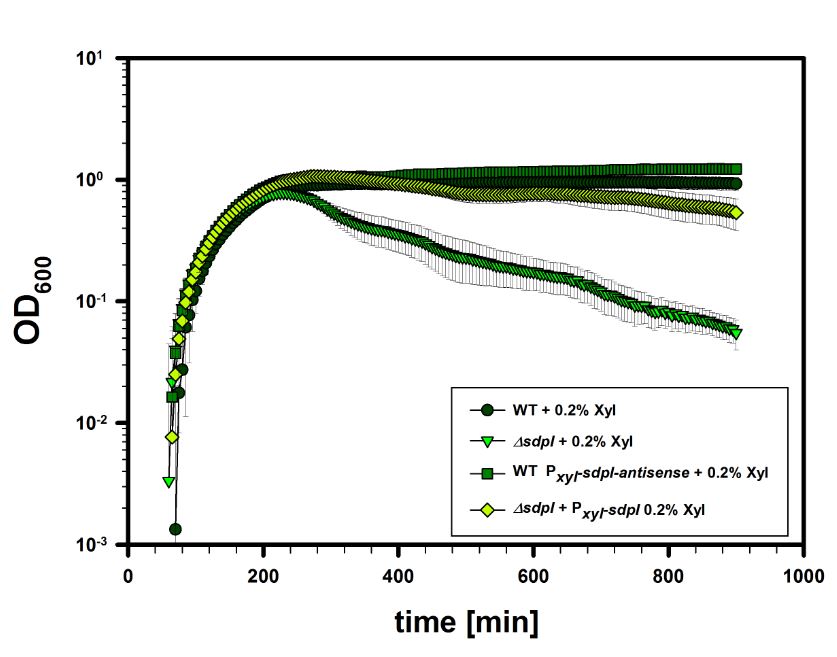
First results of this BioBrick are depicted in the following graph: | First results of this BioBrick are depicted in the following graph: | ||
Revision as of 16:20, 30 October 2014
Reverse complementary RNA sequence which binds the mRNA of the SdpI immunity
Artificial constructed complementary RNA which should bind the mRNA of the SdpI immunity protein (BBa_K1351017) in Bacillus subtilis in order to disturb the translation of immunity. Expected was a loss of immunity in B. subtilis against its own cannibalism toxin SDP (BBa_K1351043) over time.
This part was generated with the RFC10 standard and has the following prefix and suffix:
| prefix with EcoRI, NotI and XbaI: | GAATTCGCGGCCGCTTCTAGAG |
| suffix with SpeI, NotI and PstI: | TACTAGTAGCGGCCGCTGCAG |
Sites of restriction enzymes generating compatible overhangs have the same color: EcoRI and PstI in blue, NotI in green, XbaI and SpeI in red.
This part is used in the 2014 LMU-Munich iGEM project [http://2014.igem.org/Team:LMU-Munich BaKillus].
Results: [http://2014.igem.org/Team:LMU-Munich/Results LMU-2014-Results]
First results of this BioBrick are depicted in the following graph:
The graph shows grwoth curves of Bacillus subtilis wildtype, a SdpI deletion strain, a strain with our BioBrick BBa_K1351017 as well as a strain carrying our BioBrick BBa_K1351019 in trans.
The results were performed with our protocoll: Lumi Assay in MCSE and consist of three replicas of three measurements. The induction of was performed with 0.2% xylose.
These results show that our BioBrick BBa_K1351019 has no impact on the SdpI immunity. Expected was a decrease of immunity in late stationary phase somehow comparible with the deletion of SdpI.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]