Difference between revisions of "Part:BBa K1510409"
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1510409 short</partinfo> | <partinfo>BBa_K1510409 short</partinfo> | ||
A fusion of truncated INPNC anchor protein and C16 Streptococcus Mutants signal peptide becomes a bridge between E.coli and Streptococcus Mutants. This double-unit complex may not only provide a higher targeting specificity but also allow us to improve several cell-cell interactions, such as biofilm degrading and cell-holing. And in our cases, the cell-cell binding between our E.coli and Streptococcus Mutants may help us co-culture into the pathogenic-biofilm. The biofilm has long been accused of the most troublesome concrete barrier to surpass in organisms. The physical feature of biofilm often nests several pathogens, and its polysaccharide structure may serve as a safe shelter, preventing us from ridding problems from the root. However, we are flipping the game this time. With the aid of K1510816, our E.coli can attach on the Streptococcus Mutants, and be co-cultured into biofilm, giving us the chance to challenge the unsolved oral-biofilm problem. | A fusion of truncated INPNC anchor protein and C16 Streptococcus Mutants signal peptide becomes a bridge between E.coli and Streptococcus Mutants. This double-unit complex may not only provide a higher targeting specificity but also allow us to improve several cell-cell interactions, such as biofilm degrading and cell-holing. And in our cases, the cell-cell binding between our E.coli and Streptococcus Mutants may help us co-culture into the pathogenic-biofilm. The biofilm has long been accused of the most troublesome concrete barrier to surpass in organisms. The physical feature of biofilm often nests several pathogens, and its polysaccharide structure may serve as a safe shelter, preventing us from ridding problems from the root. However, we are flipping the game this time. With the aid of K1510816, our E.coli can attach on the Streptococcus Mutants, and be co-cultured into biofilm, giving us the chance to challenge the unsolved oral-biofilm problem. | ||
| + | |||
| + | First test | ||
| + | After centrifugation, the supernatant is clear; we could indicate that CSP16 is not secreted out of cell. Also, indicated from the red fluorescence of the pellet, we could say that INPNC protein display CSP16 and RFP on the surface. | ||
| + | [[file:NYMU_Cohesion picture.jpg]] | ||
| + | |||
| + | |||
| + | Second test | ||
| + | At the beginning, the population of E. coli is small as well as OD, so initial figure (Fluorescence /OD) is large. As time proceeds, Fluorescence /OD level increases dramatically, indicating that the INPNC protein is being greatly expressed. The OD level reaches a plateau at the end.[[file:NYMU_Cohesion picture2.jpg]] | ||
| + | |||
| + | |||
| + | Fluorescence level /OD level indicates the display function of INPNC through growth time.[[file:NYMU_Cohesion picture3.jpg]] | ||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 21:04, 28 October 2014
A Steptococcus Mutants adhesive complex.
A fusion of truncated INPNC anchor protein and C16 Streptococcus Mutants signal peptide becomes a bridge between E.coli and Streptococcus Mutants. This double-unit complex may not only provide a higher targeting specificity but also allow us to improve several cell-cell interactions, such as biofilm degrading and cell-holing. And in our cases, the cell-cell binding between our E.coli and Streptococcus Mutants may help us co-culture into the pathogenic-biofilm. The biofilm has long been accused of the most troublesome concrete barrier to surpass in organisms. The physical feature of biofilm often nests several pathogens, and its polysaccharide structure may serve as a safe shelter, preventing us from ridding problems from the root. However, we are flipping the game this time. With the aid of K1510816, our E.coli can attach on the Streptococcus Mutants, and be co-cultured into biofilm, giving us the chance to challenge the unsolved oral-biofilm problem.
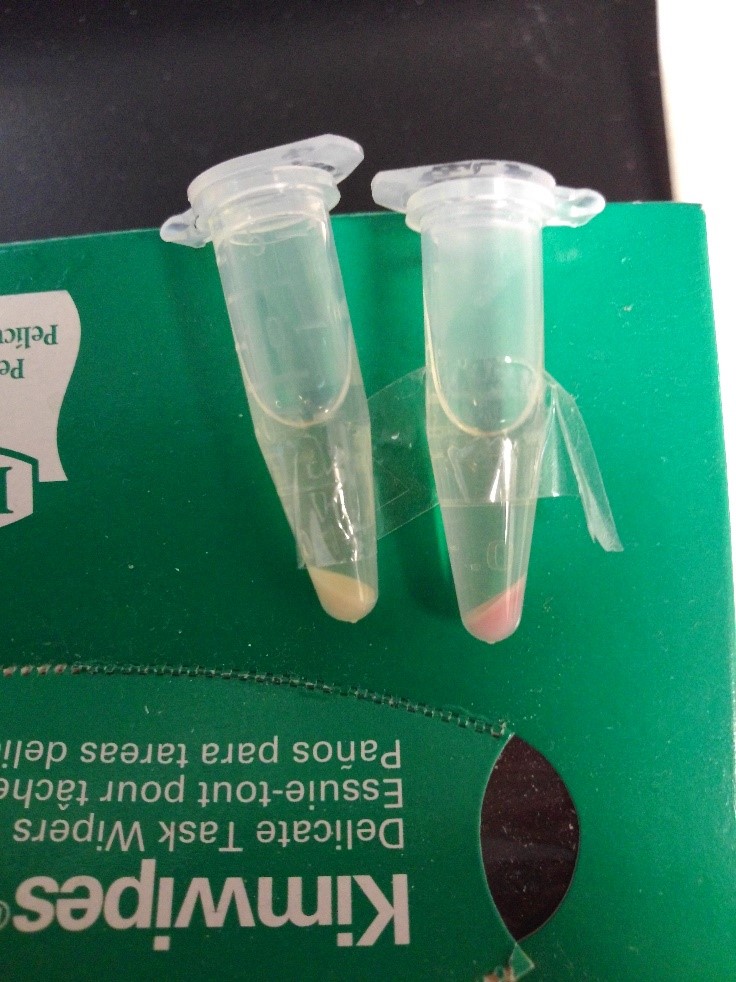
First test
After centrifugation, the supernatant is clear; we could indicate that CSP16 is not secreted out of cell. Also, indicated from the red fluorescence of the pellet, we could say that INPNC protein display CSP16 and RFP on the surface.
Second test
At the beginning, the population of E. coli is small as well as OD, so initial figure (Fluorescence /OD) is large. As time proceeds, Fluorescence /OD level increases dramatically, indicating that the INPNC protein is being greatly expressed. The OD level reaches a plateau at the end.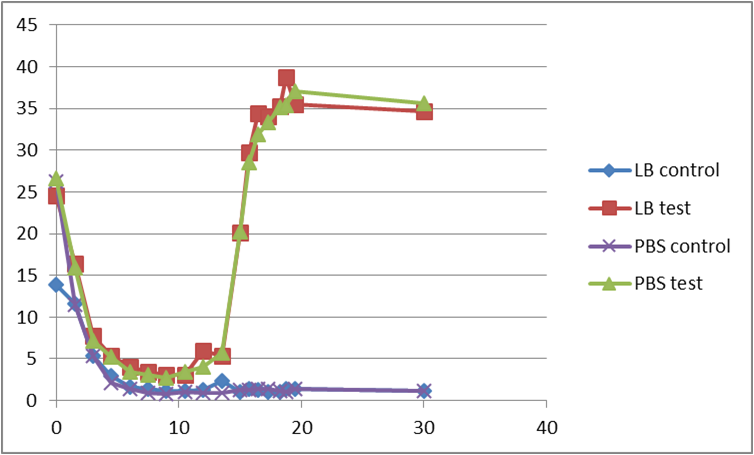
Fluorescence level /OD level indicates the display function of INPNC through growth time.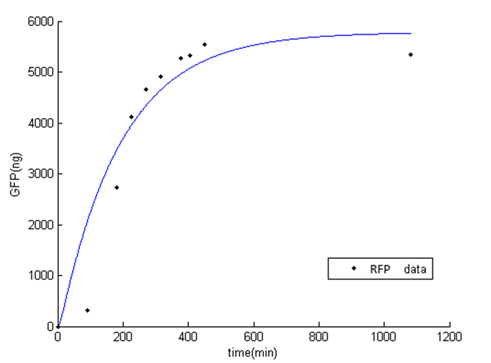
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1036
- 1000COMPATIBLE WITH RFC[1000]
