Difference between revisions of "Part:BBa J58105"
| Line 29: | Line 29: | ||
Our sensor device has been inspired on Hellinga’s work [3] sensing TNT and other molecules using a mutated periplasmic binding protein (PBP) [3]. | Our sensor device has been inspired on Hellinga’s work [3] sensing TNT and other molecules using a mutated periplasmic binding protein (PBP) [3]. | ||
| + | |||
| + | '''References''' | ||
| + | [1] [http://www.intertech.upv.es/wiki/index.php?title=Valencia_iGEM wiki Valencia team] | ||
| + | |||
| + | [2] A. Jaramillo, L. Wernisch, S. Hery and S.J. Wodak. Folding free energy function selects native-like protein sequences in the core but not on the surface. Proc. Natl. Acad. Sci. 99 (2002), 13554-13559. | ||
| + | |||
| + | [3] L.L. Looger, M.A. Dwyer, J. Smith, H.W. Hellinga. Computational design of receptor and sensor proteins with novel functions. Nature, 423, 185-190 (2003). | ||
| + | |||
| + | [4]Baumgartner J. W., Changhoon Kim, Brissette R. E., Inouye M., Chankyu Park, Hazelbauer G. L.,Journal of bacteriology 1994, vol. 176, no4, pp. 1157-1163 | ||
| + | |||
| + | [5] J.K. Joung, D.M. Koepp, and A. Hochschild. Synergistic activation of transcription by bacteriophage lambda cI protein and E. coli cAMP receptor protein. Science, 265, 1863-1866 (1994). | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
Revision as of 11:22, 29 October 2006
Synthetic periplasmic binding protein that docks a vanillin molecule
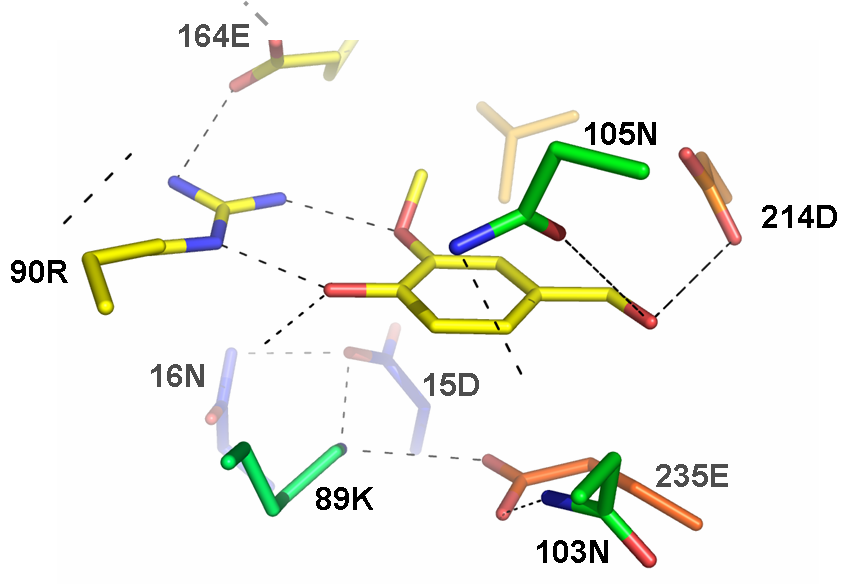
This part corresponds to the coding sequence of a computationally engineered protein designed by Pablo Tortosa and Alfonso Jaramillo at the [http://www.enseignement.polytechnique.fr/profs/biochimie/Alfonso.Jaramillo/ Synthetic Biology group at the Ecole Polytechnique (France)]. This protein is based on a ribose-binding protein template and has been designed to stabilize its closed conformation after binding a vanillin molecule. The closed conformation is able to bind a Trg protein. This part can be used to engineer a sensor genetic device if used with the part BBa_J58104 (which implements a synthetic two-component signal transduction pathway based on a Trg-EnvZ fusion protein). This device would use phosphorylated OmpR as output.
This receptor protein has been designed computationally using a ribose binding protein as a scaffold and finding the necessary mutations to change its ligand affinity towards a vanillin molecule making E. Coli tasting flavors.
By Valencia iGEM 2006
A. Aparici (2), MC. Aroca (2), J. Carrera (1), C. Edo (1,2), G. Fuertes (2), D. Jimenez (2), C. Mata (2), JV. Medrano (2), A. Montagud (2), C. Navarrete (2), E. Navarro (1), G. Rodrigo (1), M. Baguena (1), P. Tortosa (3), P. Fernandez-de-Cordoba (1), A. Ferrando (2), J. Salgado (2), J. Urchueguia (1), A. Jaramillo (3).
1 Universidad Politecnica de Valencia, Spain
2 Universitat de Valencia, Spain
3 Ecole Polytechnique, France
Usage and Biology
We can use this part to construct a sensor device. It requires using E. coli as cellular chassis (with a deficient EnvZ strain such as BBa_V1012) and the synthetic two-component signal transduction pathway implemented in part BBa_J58104. The device would then produce OmpR-P after sensing a vanillin molecule in the periplasm.
The receptor protein has been designed computationally using a ribose binding protein as a scaffold and finding the necessary mutations to change its ligand affinity towards a vanillin molecule making E. Coli tasting flavors. The computational methodology is general and can be used to sense other molecules.
When the PBP-vanillin complex binds the Trg segment of our signal transduction part BBa_J58104, an allosteric motion is propagated to the cytoplasmic EnvZ kinase domain of our chimeric protein resulting in autophosphorylation and phosphate transfer to OmpR transcription factor (OmpR-P), which is then able to induce transcription of the pOmpR promoter.
Our sensor device has been inspired on Hellinga’s work [3] sensing TNT and other molecules using a mutated periplasmic binding protein (PBP) [3].
References [1] [http://www.intertech.upv.es/wiki/index.php?title=Valencia_iGEM wiki Valencia team]
[2] A. Jaramillo, L. Wernisch, S. Hery and S.J. Wodak. Folding free energy function selects native-like protein sequences in the core but not on the surface. Proc. Natl. Acad. Sci. 99 (2002), 13554-13559.
[3] L.L. Looger, M.A. Dwyer, J. Smith, H.W. Hellinga. Computational design of receptor and sensor proteins with novel functions. Nature, 423, 185-190 (2003).
[4]Baumgartner J. W., Changhoon Kim, Brissette R. E., Inouye M., Chankyu Park, Hazelbauer G. L.,Journal of bacteriology 1994, vol. 176, no4, pp. 1157-1163
[5] J.K. Joung, D.M. Koepp, and A. Hochschild. Synergistic activation of transcription by bacteriophage lambda cI protein and E. coli cAMP receptor protein. Science, 265, 1863-1866 (1994).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Designed sequence (14 mutations, in yellow):
mnmkklatlvsavalsatvsanamaKDTIALVVETLNKPDNVSLKDGAQK
EADKLGYNLVVLDSQNNPAKELANVQDLTVRGTKILLIVPTDSDAVGNAV
KMANQANIPVITLKRQATKGEVVSHIAADNVLGGKIAGDYIAKKAGEGAK
VIELQGKAGTSAARELGEGFQQAVAAHKFNVLASQPADEDRIKGLNVMQN
LLTAHPDVQAVFAQQDEMALGALRALQTAGKSDVMVVGDVGTPDGEKAVN
DGKLAATIAELPDQIGAKGVETADKVLKGEKVQAKYPVDLKLVVKQ