Difference between revisions of "Part:BBa K1073022:Experience"
m (→Applications of BBa_K1073022) |
m (→Part Sequence Quality) |
||
| Line 10: | Line 10: | ||
===Part Sequence Quality=== | ===Part Sequence Quality=== | ||
| − | This part's real sequence may end with two stop codons, TAATAA. This is not listed in [the original source part's sequence https://parts.igem.org/Part:BBa_K592012] or in this part's sequence, but [https://parts.igem.org/cgi/sequencing/one_blast.cgi?update_database=1&id=19622 sequencing from the registry] indicates it may be present. Beware. | + | '''The Tech Museum, May 2014''': This part's real sequence may end with two stop codons, TAATAA. This is not listed in [the original source part's sequence https://parts.igem.org/Part:BBa_K592012] or in this part's sequence, but [https://parts.igem.org/cgi/sequencing/one_blast.cgi?update_database=1&id=19622 sequencing from the registry] indicates it may be present. Beware. |
===User Reviews=== | ===User Reviews=== | ||
Revision as of 00:58, 31 May 2014
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K1073022
The iGEM Team Braunschweig 2013 used this device as a reporter to distinguish two different strains when cultivated in a mixed culture.
When eforRed is expressed, the colonies on agar plates as well as the liquid culture exhibit a bright pink color.
Part Sequence Quality
The Tech Museum, May 2014: This part's real sequence may end with two stop codons, TAATAA. This is not listed in [the original source part's sequence https://parts.igem.org/Part:BBa_K592012] or in this part's sequence, but sequencing from the registry indicates it may be present. Beware.
User Reviews
UNIQ959637fc34a188fb-partinfo-00000000-QINU
|
•••••
iGEM Team Braunschweig 2013 |
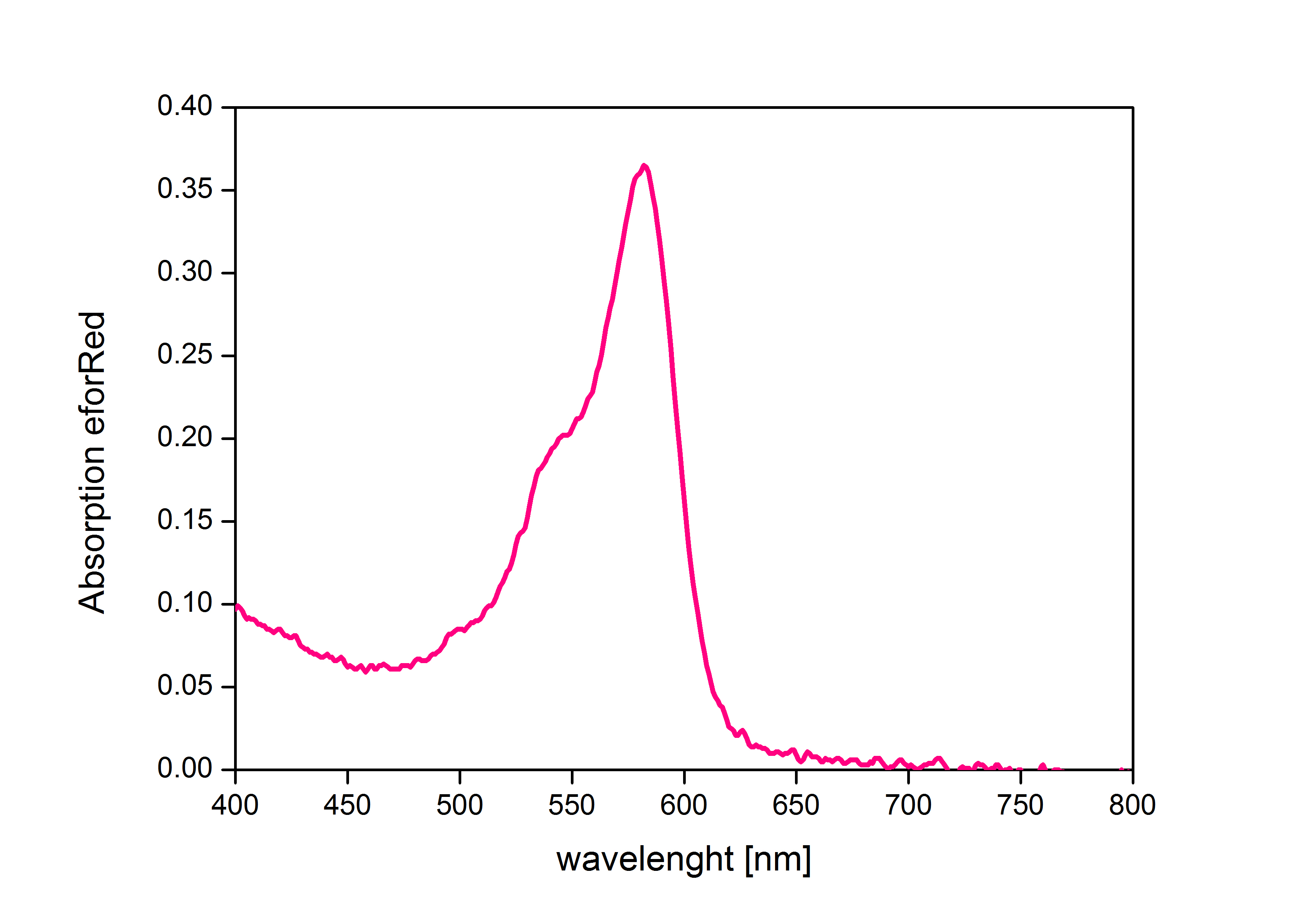
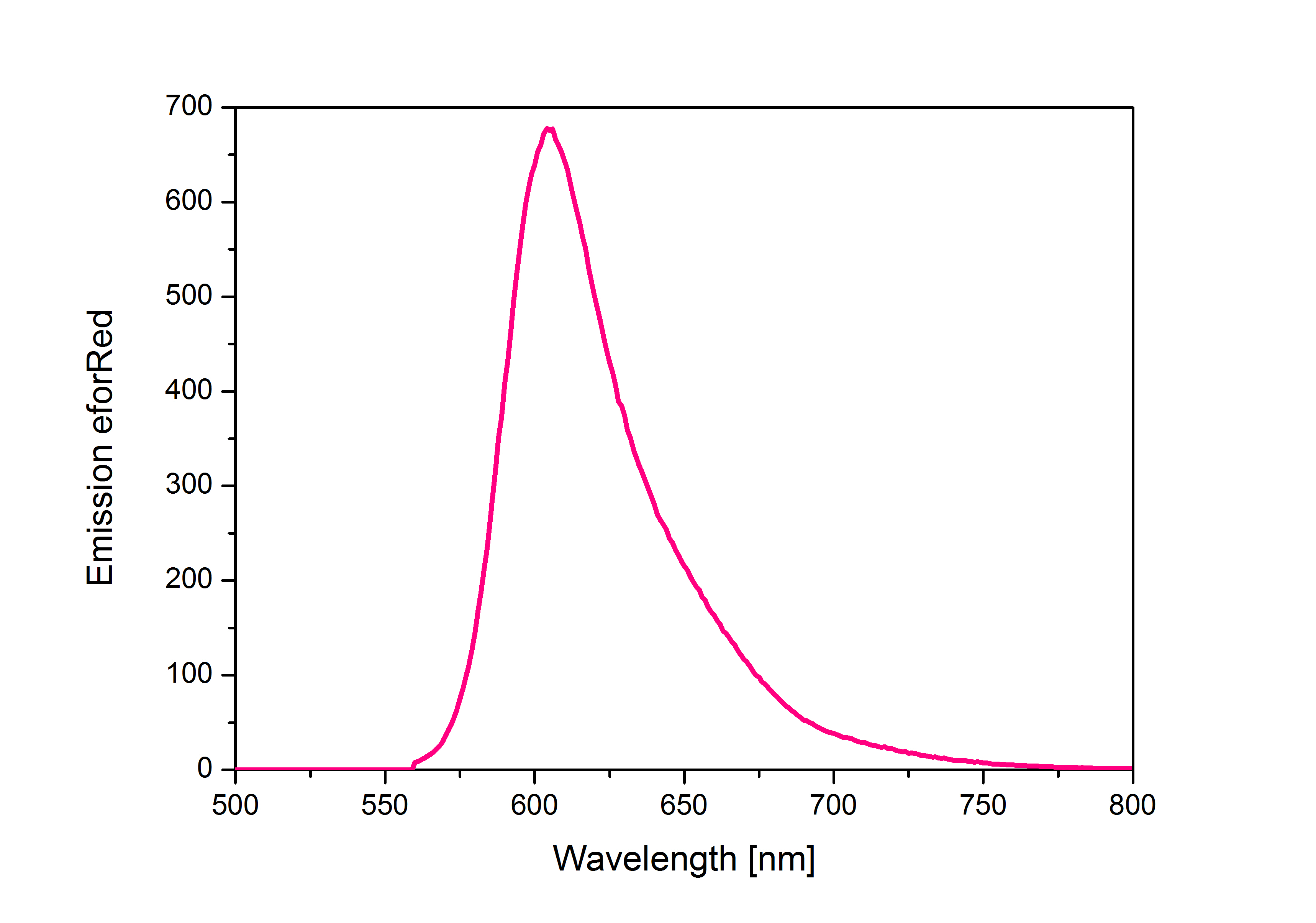
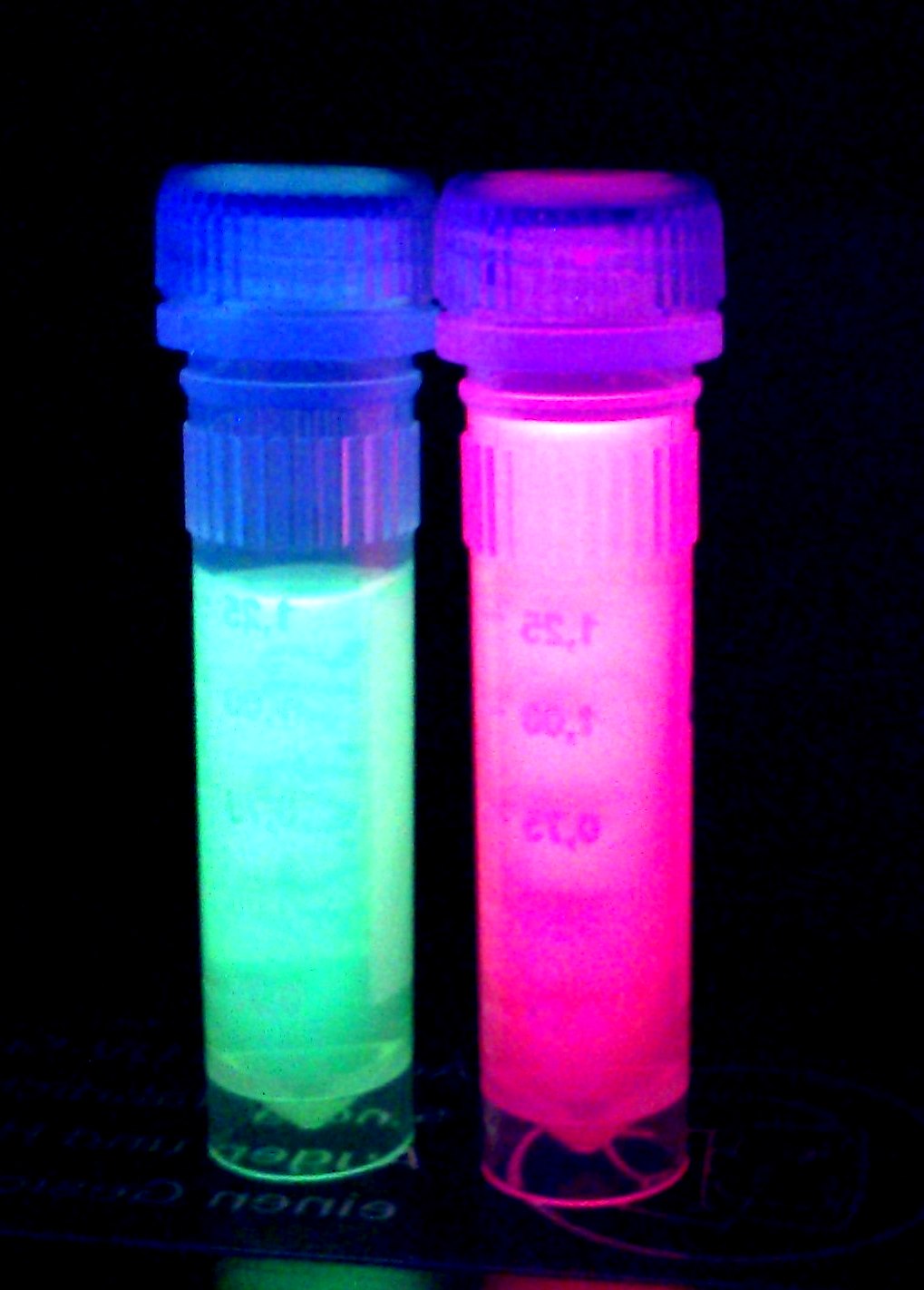
When expressed, the chromoprotein eforRed exhibits a distinct pink color. In order to avoid absorption by the chomoprotein during OD measurement of a bacterial culture, the absorption and emission spectra were measured. An optimal wavelength for spectral optical density measurement was determined to be at 520nm. iGEM Team Braunschweig 2013: Absorptionspectrum of chromoprotein eforRed in the wavelength range 400-800 nm. Absorption maximum of eforRed: 534 nm iGEM Team Braunschweig 2013: Emission spectrum of chromoprotein eforRed. The chromoprotein does show significant emission and can be detected by its fluorescence. Emission maximum of eforRed: 605 nm iGEM Team Braunschweig 2013: Supernatant after cell disruption containing chromoproteins of eforRed and amilGFP (BBa_K1073024). Both chromoproteins fluoresce when excited by UV-light. |
UNIQ959637fc34a188fb-partinfo-00000003-QINU