Difference between revisions of "Part:BBa K1216008"
| Line 4: | Line 4: | ||
BBa_K1216008: part of a P<sub>LuxR</sub> collection of promoters with differing AHL sensitivity. | BBa_K1216008: part of a P<sub>LuxR</sub> collection of promoters with differing AHL sensitivity. | ||
This significantly increases usefulness of the Lux signaling system, as differential respones to cell concentrations are enabled. Further they can even be used to detect geometrical settings of colonies. | This significantly increases usefulness of the Lux signaling system, as differential respones to cell concentrations are enabled. Further they can even be used to detect geometrical settings of colonies. | ||
| + | |||
| + | After obtaining a [https://parts.igem.org/Part:BBa_K1216007 less sensitive variant] of [https://parts.igem.org/Part:BBa_R0062 P<sub>LuxR</sub>] we used it as a starting point for [https://parts.igem.org/Part:BBa_K1216008:Design further engineering] of mutant promoters. This approach ultimately lead to a library of promoters spanning a reasonably big range of EC<sub>50</sub> values. Please see details in the [https://parts.igem.org/Part:BBa_K1216008#Characterization characterization section] below or by clicking the characterization tab. | ||
<br clear="all"> | <br clear="all"> | ||
<!-- Add more about the biology of this part here--> | <!-- Add more about the biology of this part here--> | ||
Revision as of 14:00, 30 October 2013
Variant of the wild-type pLuxR promoter with lower sensitivity
BBa_K1216008: part of a PLuxR collection of promoters with differing AHL sensitivity. This significantly increases usefulness of the Lux signaling system, as differential respones to cell concentrations are enabled. Further they can even be used to detect geometrical settings of colonies.
After obtaining a less sensitive variant of PLuxR we used it as a starting point for further engineering of mutant promoters. This approach ultimately lead to a library of promoters spanning a reasonably big range of EC50 values. Please see details in the characterization section below or by clicking the characterization tab.
Usage and Biology

The Lux signaling system is based on positive feedback. The PLuxL promoter drives expression of the upstream luxR gene. The promoter PLuxR driver expression of the downstream operon including the LuxI AHL synthetase. The promoter encompasses a palindromic operator sequence to which a dimer of LuxR-AHL complexes binds to induce transcription. In absence of this complex, PLuxL is active and thus LuxR is abundant. PLuxR however is inactive, showing slight leakage, yielding a fairly low expression level of LuxI and other downstream gene products.
An important feature of AHL, the "signaling molecule" of the system, is that it diffuses freely through cell walls and surrounding media. This enables communication between cells in vincinity. When a certain cell concentration is reached, the diffusing AHL yields sufficiently high levels of the LuxR-AHL complex within cells. The direct consequence of this is high expression of downstream genes, importantly inculding LuxI (and not so importantly shutting down PLuxL). Now the positive feedback loop is being closed by elevated synthesis of AHL which further increases LuxI synthesis in cells.
In V. Fischeri other downstream genes include a luciferase which is part of a symbiosis with marine animals. Other possibilities are proteins that facilitate biofilm formation and related phenomena.
In (synthetic) biology, this system can be used as a sender/reporter system by separating LuxI and LuxR, where LuxI is not under control of PLuxR, rendering the system independent of cell concentration. LuxR/PLuxR can then be used to drive expression of a myriad of other genes such as various reporter genes.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
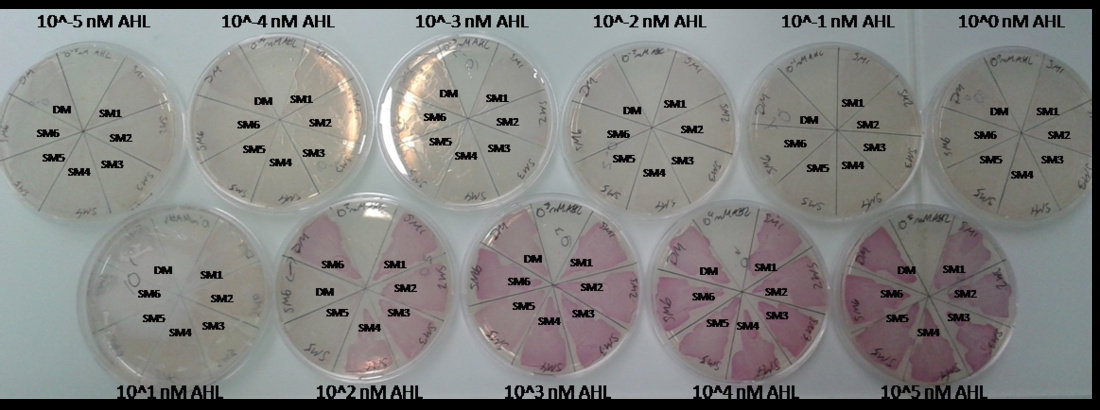
As explained above we created a library of promoters originating from PLuxR variant (G1). We characterized the dose response curve to AHL on agar plates of all 7 promoters by following the same protocol as for the PLuxR variant characterization. See the different shifted dose response curves in Figure 10. and the characteristics of the library in Table 1.
Thanks to the analytical solution of the model (see [http://2013.igem.org/Team:ETH_Zurich/Modeling/Analytical_Approximations here]) predicting the needed EC50 of 100 nM of the promoter to fit to our system we choose the SM1 mutant for our final set-up. (We did not choose SM2 because when we choose a promoter, according to the prediction of the model, the experiments were not done in duplicates at this time, and the characteristics changed of course as soon as we had duplicates). This promoter gets activated when two mines are nearby the receiver cell.

Figure 10. shows the relative fluorescence over the [AHL] of the dose-response curves of the whole library exepted the G1 and the wild type PLuxR promoter. Figure 11. is a comparison of a choosen set of promissing promoter we tried in different constructs for the final gameplay according to the prediction of the model.
Fluorescence data analysis
The fitting of the following graphs was performed using this equation :
Output = eGFP levels [au]
Top = maximal eGFP level [au] ("full induction")
Bottom = minimal eGFP level [au] (“leakiness”)
n = Hill coefficient (“cooperativity”)
EC50 = Half-maximal effective concentration (“sensitivity”)
[AHL]=AHL concentration [nM]
References



