Difference between revisions of "Part:BBa K1152009"
m |
|||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1152009 short</partinfo> | <partinfo>BBa_K1152009 short</partinfo> | ||
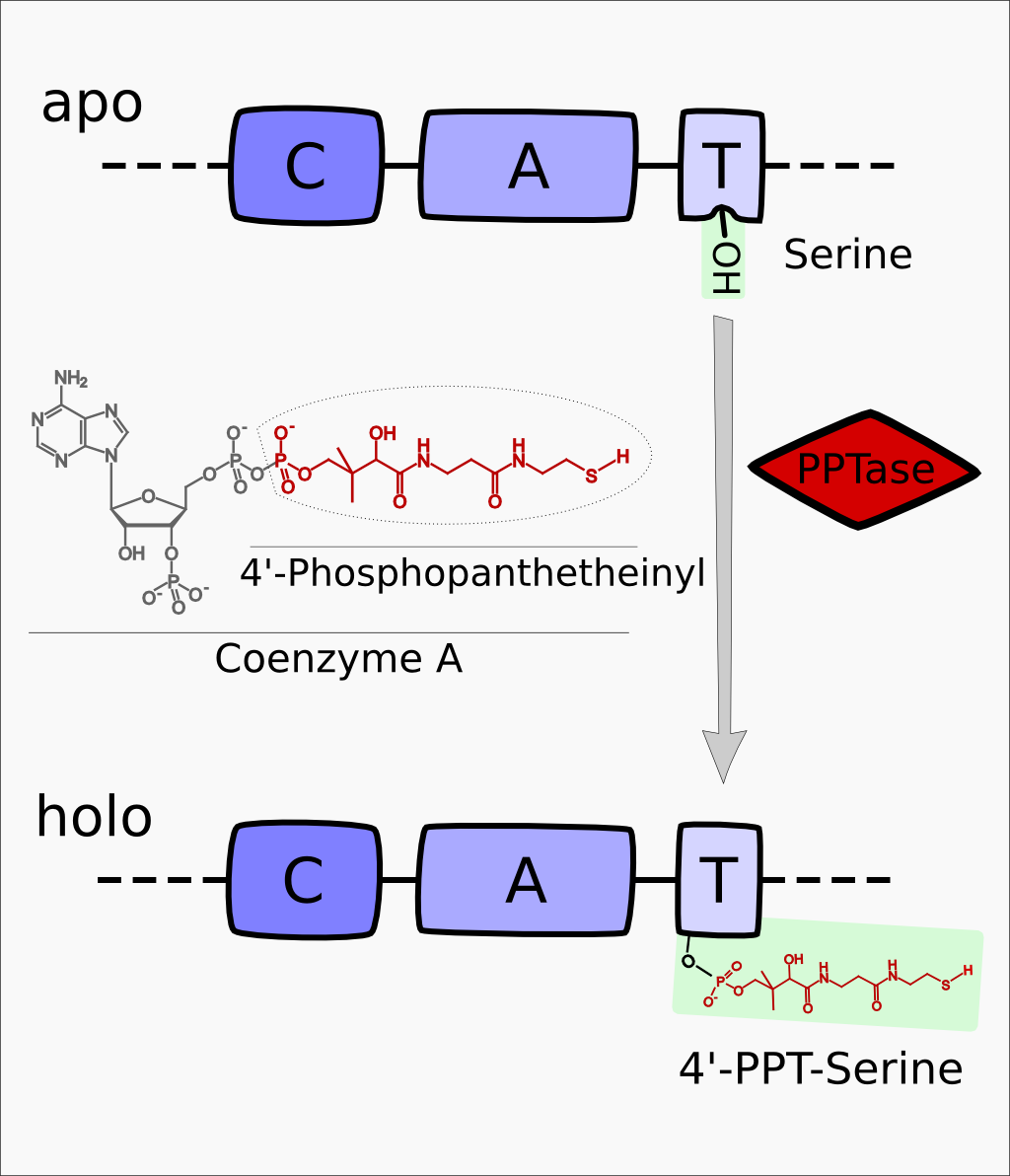
| − | + | Sfp encodes for a 4'-Phosphopanthetheinyl-transferase (PPTase) which transfers a 4'-PPT residue from CoA to a conserved serine residue in the T-Domain of NRPS modules, thus activating these enzymes. | |
| − | transfers a 4'-PPT residue from CoA to a conserved serine residue in the | + | |
| − | T-Domain of NRPS modules, thus activating these enzymes. | + | |
<gallery> | <gallery> | ||
| − | + | File:Heidelberg2013_PPtase_mechanism.png|'''Figure 1:''' General mechanism of activation the activation of NRP Synthetases from apo- to holo-form. | |
</gallery> | </gallery> | ||
| − | We hereby improve parts <partinfo>BBa_K802006</partinfo> and | + | We hereby improve parts <partinfo>BBa_K802006</partinfo> and <partinfo>BBa_K302010</partinfo> which also encode for sfp from ''Bacillus subtilis''. Both parts are equal in sequence dispite the fact that obviesly <partinfo>BBa_K802006</partinfo> includes additional bases upstream from the ''B. subtilis'' genome. we improved these parts in availablity to the community as well as in characterization. |
| − | <partinfo>BBa_K302010</partinfo> which also encode for sfp from | + | |
| − | ''Bacillus subtilis''. Both parts are equal in sequence dispite the fact | + | |
| − | that obviesly <partinfo>BBa_K802006</partinfo> includes additional bases | + | |
| − | upstream from the ''B. subtilis'' genome. we improved these parts in | + | |
| − | availablity to the community as well as in characterization. | + | |
| − | By MSA we can conclude that <partinfo>BBa_K1152009</partinfo> is | + | By MSA we can conclude that <partinfo>BBa_K1152009</partinfo> is different from <partinfo>BBa_K802006</partinfo> and <partinfo>BBa_K302010</partinfo> at 3 different amino acid sites: [[File:Heidelberg2013_BBa_sfp_comparement.clustal.txt|MSA sfp BioBricks]]. |
| − | different from <partinfo>BBa_K802006</partinfo> and | + | |
| − | <partinfo>BBa_K302010</partinfo> at 3 different amino acid sites: | + | |
| − | [[File:Heidelberg2013_BBa_sfp_comparement.clustal|MSA sfp BioBricks]]. | + | |
| − | We can also conclude that <partinfo>BBa_K802006</partinfo> is actually | + | We can also conclude that <partinfo>BBa_K802006</partinfo> is actually sfp even when not annotated like this. |
| − | sfp even when not annotated like this. | + | |
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | Sfp is a unspecific PPTase widly used for activating NRPSs. [Lambalot, 1996] | |
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
| Line 33: | Line 22: | ||
| − | <!-- Uncomment this to enable Functional Parameter display | + | <!-- Uncomment this to enable Functional Parameter display |
===Functional Parameters=== | ===Functional Parameters=== | ||
<partinfo>BBa_K1152009 parameters</partinfo> | <partinfo>BBa_K1152009 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
Revision as of 22:54, 5 October 2013
Sfp 4'-Phosphopanthetheinyl-transferase
Sfp encodes for a 4'-Phosphopanthetheinyl-transferase (PPTase) which transfers a 4'-PPT residue from CoA to a conserved serine residue in the T-Domain of NRPS modules, thus activating these enzymes.
We hereby improve parts BBa_K802006 and BBa_K302010 which also encode for sfp from Bacillus subtilis. Both parts are equal in sequence dispite the fact that obviesly BBa_K802006 includes additional bases upstream from the B. subtilis genome. we improved these parts in availablity to the community as well as in characterization.
By MSA we can conclude that BBa_K1152009 is different from BBa_K802006 and BBa_K302010 at 3 different amino acid sites: File:Heidelberg2013 BBa sfp comparement.clustal.txt.
We can also conclude that BBa_K802006 is actually sfp even when not annotated like this.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 651
Illegal SapI.rc site found at 661