Difference between revisions of "Part:BBa K1152006"
(→Verification and Biology) |
(→Verification and Biology) |
||
| Line 17: | Line 17: | ||
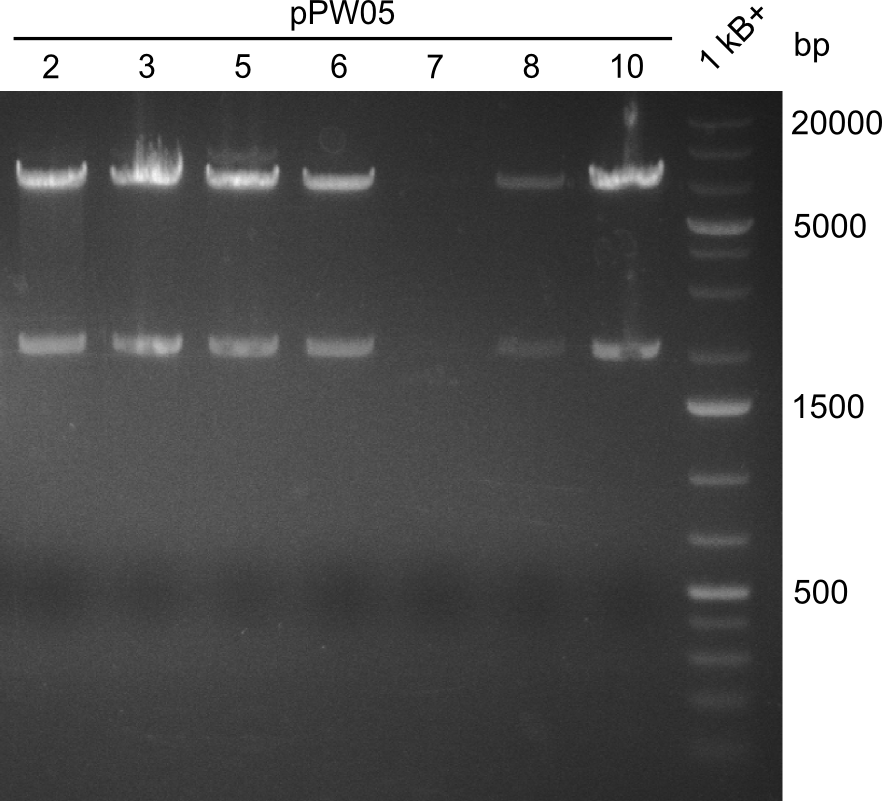
EcoRI and PstI, as expected two bands show up, one at around 2000 bp (backbone) and one at around 7000 bp (insert). | EcoRI and PstI, as expected two bands show up, one at around 2000 bp (backbone) and one at around 7000 bp (insert). | ||
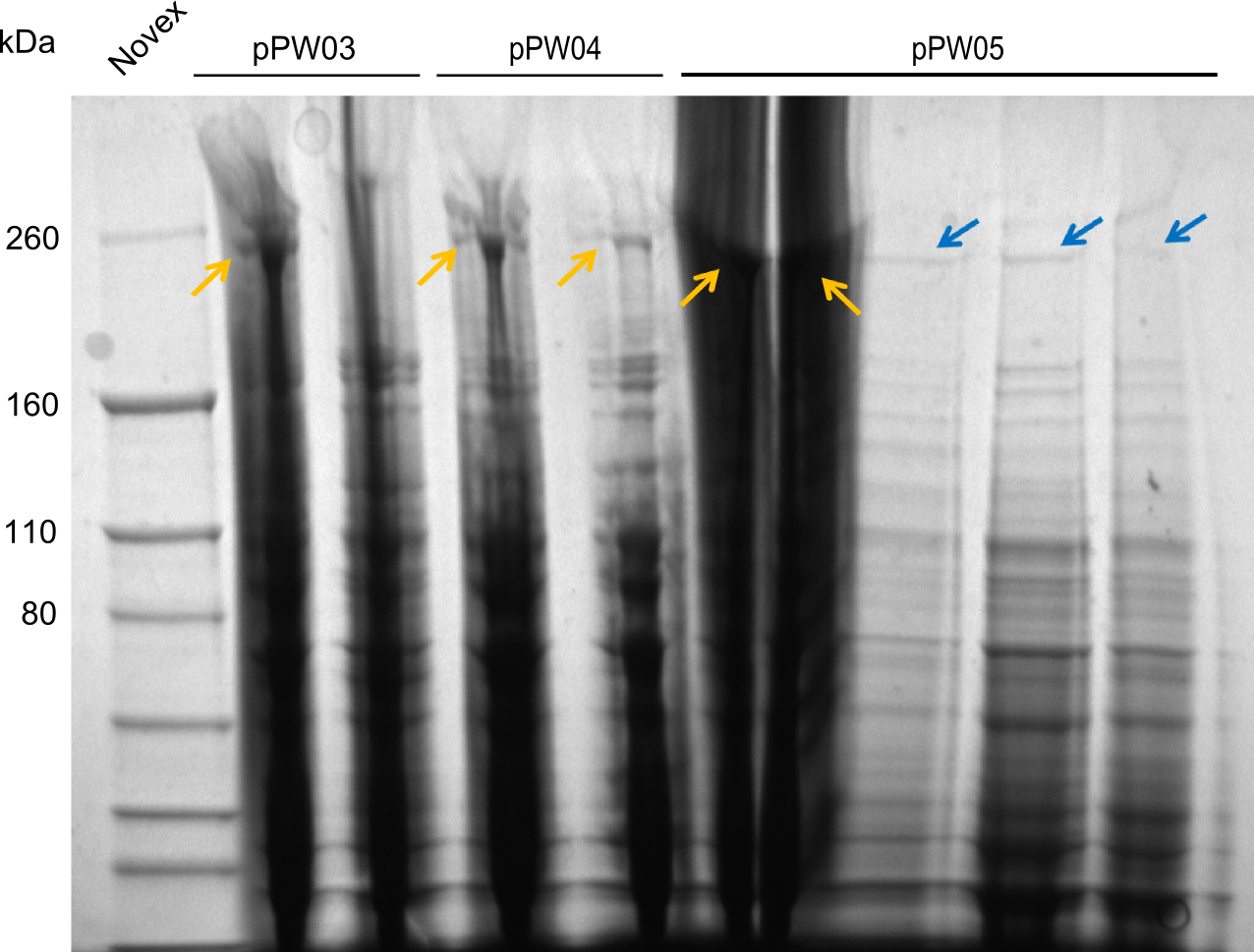
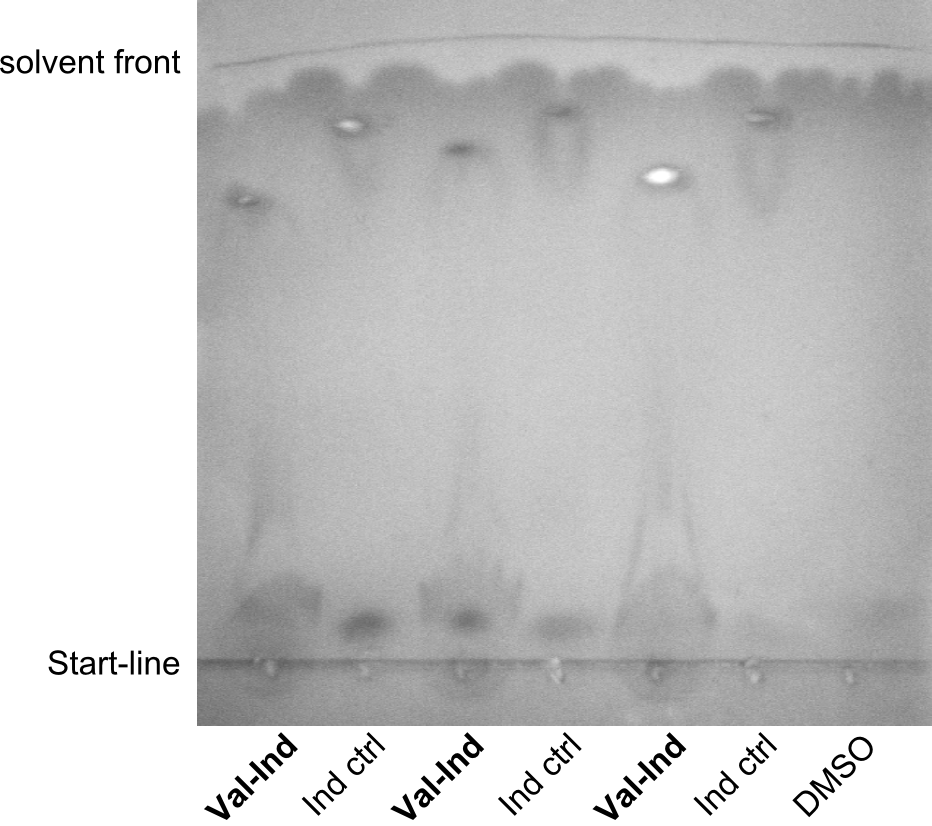
| − | In the SDS-PAGE, expression of the NRPS could be verified, as the expected band at 242kDa is clearly visible after induction with IPTG (see figure 2). To prove that the Val-Ind NRPS is in fact functional, we purified the synthesized NRP and analyzed its properties by comparative TLC with an Indigoidine control. As one can see in figure 3, the migration | + | In the SDS-PAGE, expression of the NRPS could be verified, as the expected band at 242kDa is clearly visible after induction with IPTG (see figure 2). To prove that the Val-Ind NRPS is in fact functional, we purified the synthesized NRP and analyzed its properties by comparative TLC with an Indigoidine control. As one can see in figure 3, the migration-length of the synthetic fusion peptide is significantly slower compared the one of Indigoidine. The samples for Val-Ind tested on TLC were extracted from different clones, hence reproducibility by biological replicates could be proven. |
Latest revision as of 02:08, 5 October 2013
Valine-Indigoidine fusion - NRPS
Non-Ribosomal Peptide Synthetase that synthesizes a fusion peptide consisting of Valine and Indigoidine, where the NRPS-modules are from the Tyc-Cluster from B. parabrevis and from the indC-gene from P. luminescens.
Verification and Biology
pPW05 was amplified out of four fragments via Gibson Cloning: The module TycC4dC, the C-domain of TycC2 and the indC-gene, as well as a backbone. We verified the genotype by colony-PCR, restriction digest with various restriction enzymes, among them EcoRI and PstI, and sequencing of the assembly overhang regions. Furthermore, SDS-PAGE was performed after induction of cells in order to prove expression – the expected band at 242 kDa was clearly visible after Coomassie staining. Finally, after purification of the NRP, TLC on silica-gel with Dichloromethane as mobile phase, was performed to prove the functionality of the expressed NRPS.
(Figures 1, 2 and 3: Verification of the genotype, the expression and the functionality of BBa_K1152006)
In restriction digest, as one can see in the first figure, the cutting pattern was as expected. For restriction digest with EcoRI and PstI, as expected two bands show up, one at around 2000 bp (backbone) and one at around 7000 bp (insert).
In the SDS-PAGE, expression of the NRPS could be verified, as the expected band at 242kDa is clearly visible after induction with IPTG (see figure 2). To prove that the Val-Ind NRPS is in fact functional, we purified the synthesized NRP and analyzed its properties by comparative TLC with an Indigoidine control. As one can see in figure 3, the migration-length of the synthetic fusion peptide is significantly slower compared the one of Indigoidine. The samples for Val-Ind tested on TLC were extracted from different clones, hence reproducibility by biological replicates could be proven.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 4118
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 962
Illegal SapI.rc site found at 1158
Illegal SapI.rc site found at 5381