Difference between revisions of "Part:BBa K1150003"
Lisaschmunk (Talk | contribs) |
|||
| Line 68: | Line 68: | ||
| − | + | ==References== | |
<small> | <small> | ||
<!-- | <!-- | ||
Revision as of 13:32, 4 October 2013
G9a
| G9a | |
|---|---|
| Function | Histone Methyltransferase |
| Use in | Mammalian cells |
| RFC standard | RFC 25 |
| Backbone | pSB1C3 |
| Organism | Mus musculus |
| Source | Albert Jeltsch, Stuttgart |
| Submitted by | [http://2013.igem.org/Team:Freiburg Freiburg 2013] |
This BioBrick is part of the Freiburg 2013 uniCAS toolkit for gene regulation and contains the histone methyltransferase (HMTase) G9a encoded by the murine EHMT2 gene. G9a is able to induce heterochromatization by transferring methyl groups to lysine 9 of histone 3 (H3K9) [1]. This part only contains the catalytically active SET domain of G9a that is responsible for histone modification. By fusing an HMTase domain to the DNA binding dCas9 device, specific endogenous gene targeting is possible. In regions of open chromatin structures H3K9 methylation can therefore lead to inactivation of the concerned locus. In combination with a customized RNAimer plasmid different loci can be targeted for transcriptional repression of gene expression.
Usage and Biology
The murine EHMT2 (Euchromatic histone-lysine N-methyltransferase 2) gene encodes a mammalian lysine-preferring histone methyltransferase called G9a [1]. The complete G9a protein contains a SET (Su(var)3-9, Enhancer of Zeste, Trithorax) domain, which catalyzes transfer of methyl groups to H3K9 and an ANK domain which has a role in de novo DNA methylation [2]. The G9a BioBrick only comprises the G9a-SET domain. To provide the possibility of stable endogenous repression in our uniCAS toolkit we constructed the following four devices using the G9a BioBrick.
- the uniCAS Histone Modifier (CMV promoter)
- the uniCAS Histone Modifier (SV40 promoter)
- the uniCAS Red Light Switch Part II - Histone Modifier is a result of combining G9a with part of our red light switch, Phytochrome B. PhyB will dimerize with PIF6 upon red light stimulus. PIF6 is fused to dCas9 in the uniCAS Red Light Switch Part I - Stimulator. Using these two devices G9a is brought into proximity of the DNA targeting dCas9 and should therefore be able to site specifically transfer methyl groups to histone H3 and to repress gene expression.
- the uniCAS UV Light Switch Part II - Histone Modifier
Expression tests
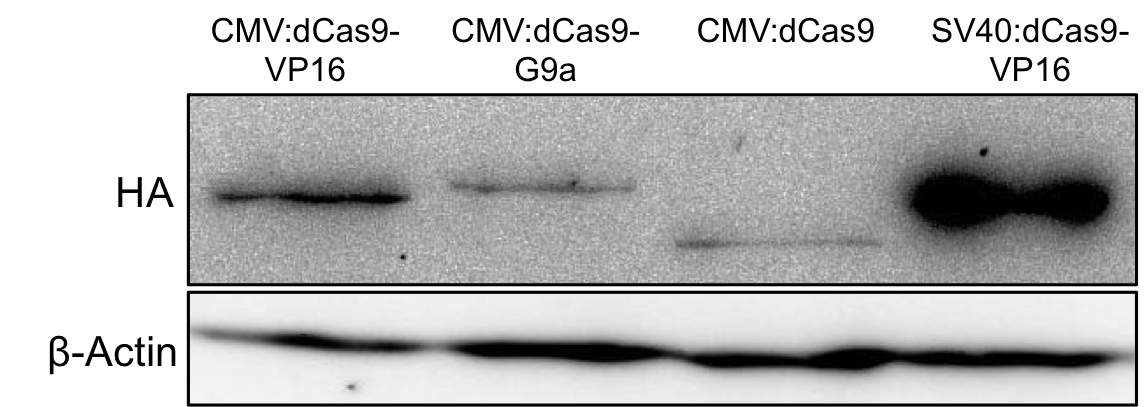
The expression of G9a fusion constructs was determined via Western blotting. Therefore mammalian HEK293T cells were transfected with the uniCAS Histone Modifier (CMV promoter) in 6-well plates (150,000 cells/well). 48h after transfection the cells were harvested and the complete cell lysate was blotted with anti-HA antibody against the N-terminally HA-tagged dCas9 proteins. The fullength dCas9-G9a protein could be detected at the expected size of approximately 185kDa.
Figure 2.: HA-tag fused to different dCas9-fusion proteins encoded on the RFC25 pSB1C3 backbone.
Functional tests
Targeting of dCas9-G9a to an open, endogenous locus should lead to changes in the epigenetic chromatin state and repression of gene expression. To test this, the endogenous VEGF-A locus was chosen, as it is well characterized and open in HEK-293T cells. Additionally, VEGF production can easily be measured by ELISA analysis.
Cells were seeded into 24-well format at a density of 65,000 cells per well and were co-transfected with the desired crRNAs, based on the RNAimer. These plasmids are derivates of the RNAimer, the only difference was the insertion of the target sites. These were: -8, -573, +434, -475 and -573/+434. As an off-target control crRNA targeting EMX was used. Every site was targeted with CMV:dCas9 wthout G9a, to differentiate the action of G9a from the CRISPRi effect. As an internal standard a constitutive SEAP reporter was used. 24 hours post transfection, VEGF levels and SEAP levels were measured and the ratio was calculated. Error bars represent standard deviations.
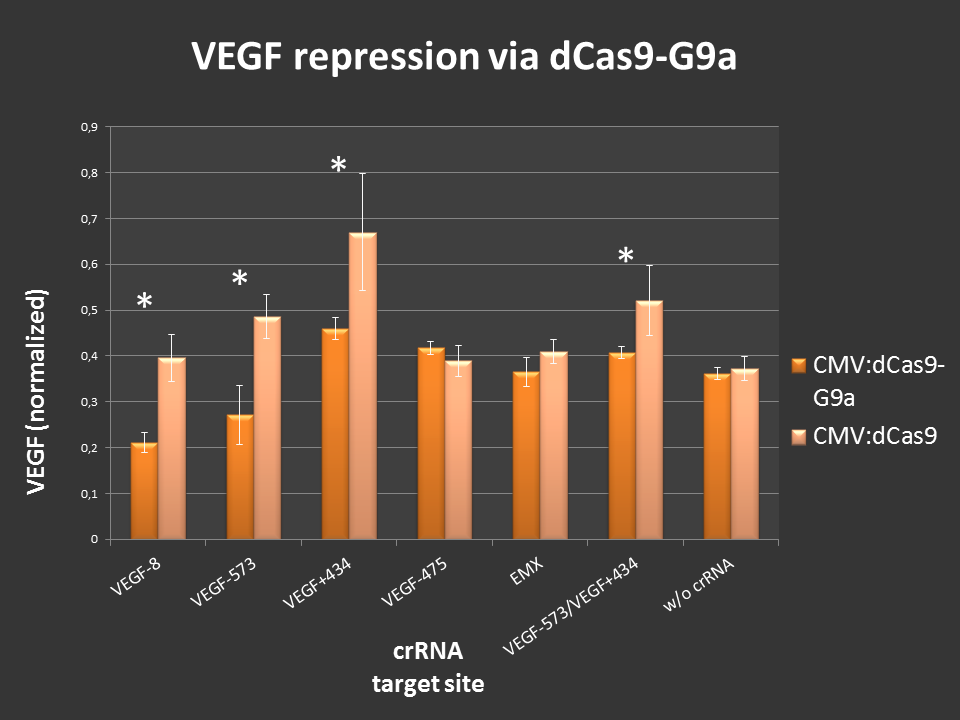
Figure 3.: dCas9-G9a targeting of endogenous VEGF-A locus in HEK-293T cells.
ELISA values, normalized to internal SEAP standard. For some loci clear repressive effects up to 50% were detectable. Controls did not display any off-target effects. Combining two target sites did not show stronger repression than targeting one site at a time.
Different VEGF target sites led to different repression efficiencies and simultaneous targeting of two sites did not further enhance the observed effects.
In summary, the CMV:dCas9-G9a is an effective repressor of endogenous gene expression, by modulating the chromatin structure of the targeted locus. This offers application in fundamental research as well as in medical sciences.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 597
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
[1] Tachibana, M., et al. (2001). SET Domain-containing Protein, G9a, Is a Novel Lysine-preferring Mammalian Histone Methyltransferase with Hyperactivity and Specific Selectivity to Lysines 9 and 27 of Histone H3. The Journal of Biological Chemistry, 276, 25309-25317.
[2] Epsztejn-Litman, S., et al. (2008). De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 15(11): 1176–1183.