Difference between revisions of "Part:BBa K818000:Experience"
(→User Reviews) |
(→User Reviews) |
||
| Line 105: | Line 105: | ||
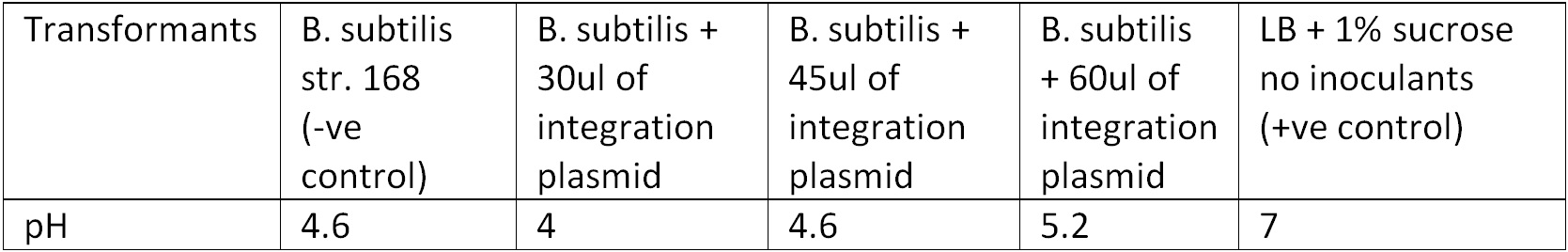
Figure 3. Display the pH of each samples following the Phenol Red Sucrose test on overnight grown culture in LB + 1% Sucrose media. | Figure 3. Display the pH of each samples following the Phenol Red Sucrose test on overnight grown culture in LB + 1% Sucrose media. | ||
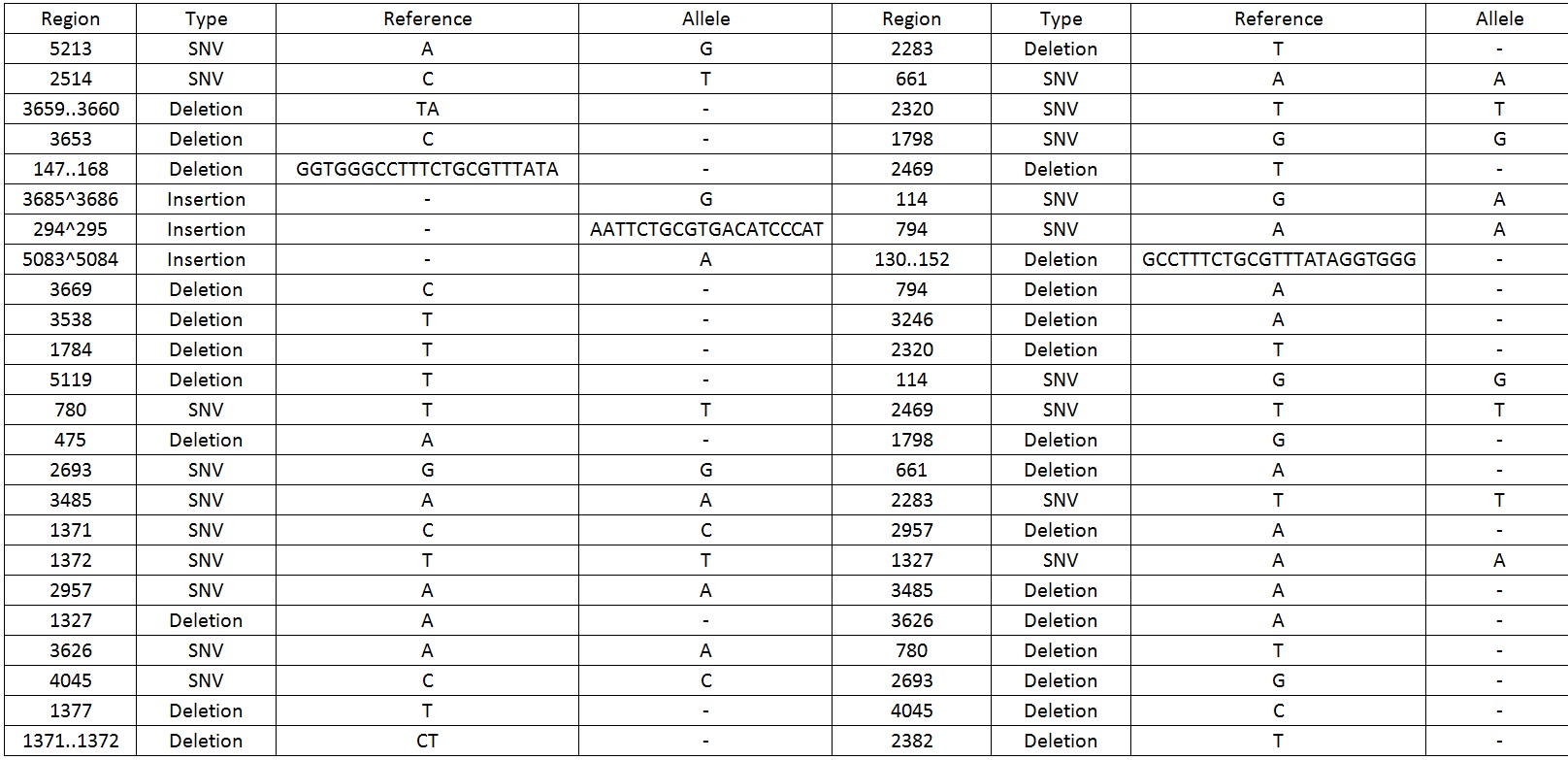
| − | We then sequenced the backbone and found out that there were 49 mutations which include SNV, deletions and insertions. | + | We then sequenced the backbone and found out that there were 49 mutations which include SNV, deletions and insertions.Table 1 display the mutations found on the sequence. |
[[File:BareCillus_Gro_mutations.jpg|700px]] | [[File:BareCillus_Gro_mutations.jpg|700px]] | ||
| − | + | Table 1. Shows all the mutations found in the BBa_K818000 backbone. | |
| − | Table 1 | + | |
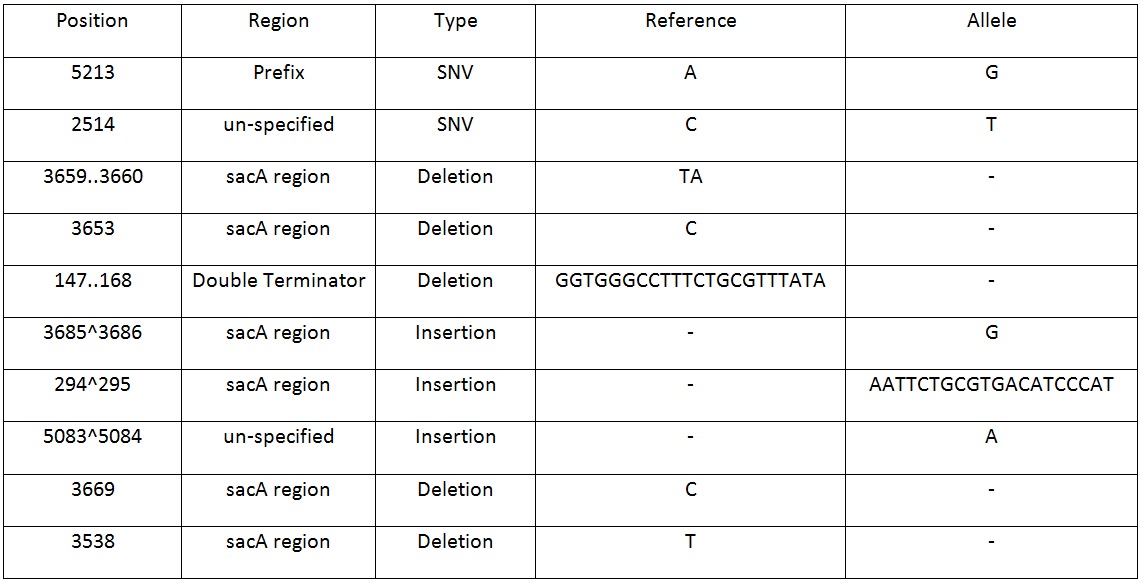
| + | Table 2. Shows list of mutations found in the BBa_K818000 backbone, including the position, region and type of mutations after analysing the initial sequencing and the sequencing of highly mutated regions. | ||
[[File:BareCillus Gro sumseq.jpg|700px]] | [[File:BareCillus Gro sumseq.jpg|700px]] | ||
| − | |||
Table 2. Shows list of mutations found in the BBa_K818000 backbone, including the position, region and type of mutations after analysing the initial sequencing and the sequencing of highly mutated regions. | Table 2. Shows list of mutations found in the BBa_K818000 backbone, including the position, region and type of mutations after analysing the initial sequencing and the sequencing of highly mutated regions. | ||
| + | |||
The results from both sequencing run proved to show similar mutations were found on this backbone, most of the mutations occurred in the sacA integration regions. These results explained the reason why this pSac-Cm derived integration backbone for ‘’B.subtilis’’ were not working. | The results from both sequencing run proved to show similar mutations were found on this backbone, most of the mutations occurred in the sacA integration regions. These results explained the reason why this pSac-Cm derived integration backbone for ‘’B.subtilis’’ were not working. | ||
Revision as of 17:11, 30 September 2013
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K818000
This backbone plasmid was used as primary backbone for all constructs in team Groningen 2012 project: the Food Warden. For further info: [http://2012.igem.org/Team:Groningen/OurBiobrick iGEM Groningen 2012 biobrick page]
User Reviews
UNIQ1dd55e32806451c5-partinfo-00000000-QINU