Difference between revisions of "Part:BBa K1092003:Experience"
(→Characterization of BBa_K1092003) |
(→Characterization of BBa_K1092003) |
||
| Line 6: | Line 6: | ||
===Characterization of BBa_K1092003=== | ===Characterization of BBa_K1092003=== | ||
1. Biobrick DNA length verification | 1. Biobrick DNA length verification | ||
| − | To check the inserted biobrick DNA length, PCR was conducted. | + | To check the inserted biobrick DNA length, PCR was conducted (Figure 1). |
[[File:di.jpg]] | [[File:di.jpg]] | ||
| Line 17: | Line 17: | ||
| − | Time series analysis were performed. The concentration of hydroquinone was monitored by UV spectrophotometer at 298 nm. According to a chi-square test (Rosenberg ''et al''., 1998), the trends of hydroquinone degradation rate in E. coli BL21 transformed with catechol 1,2-dioxygenase and that in wild type E. coli BL21 were very highly statistically different (p < 0.001) (Figure | + | Time series analysis were performed. The concentration of hydroquinone was monitored by UV spectrophotometer at 298 nm. According to a chi-square test (Rosenberg ''et al''., 1998), the trends of hydroquinone degradation rate in E. coli BL21 transformed with ''catechol 1,2-dioxygenase'' and that in wild type E. coli BL21 were very highly statistically different (p < 0.001) (Figure 2). |
Revision as of 02:32, 28 September 2013
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Characterization of BBa_K1092003
1. Biobrick DNA length verification
To check the inserted biobrick DNA length, PCR was conducted (Figure 1).
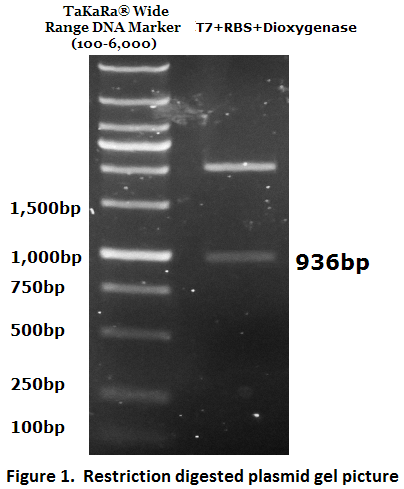
The PCR product meets the target size (936bp), the DNA sequence was further verified by DNA sequencing.
2. Protein expression verification by time series degradation assay
Catechol 1,2-dioxygenase is resposible for phenol degredation (Naiem et al., 2011). Before it is integrated into our PAHs degrdation system, hydroquinone, also known as benzene-1,4-diol or quinol, which is an aromatic organic compound belonging to the phenol family, was used as a substrate to test Catechol 1,2-dioxygenase expression and activity.
Time series analysis were performed. The concentration of hydroquinone was monitored by UV spectrophotometer at 298 nm. According to a chi-square test (Rosenberg et al., 1998), the trends of hydroquinone degradation rate in E. coli BL21 transformed with catechol 1,2-dioxygenase and that in wild type E. coli BL21 were very highly statistically different (p < 0.001) (Figure 2).
User Reviews
UNIQ9f1c7c83339638ed-partinfo-00000000-QINU UNIQ9f1c7c83339638ed-partinfo-00000001-QINU

