Difference between revisions of "Part:BBa K1100014"
(→Background) |
(→Background) |
||
| Line 25: | Line 25: | ||
Riboswitches are regulatory RNAs that regulate gene expression by binding small molecules. Recently, a novel riboswitch was reported which is present in the leader RNA of the aminoglycoside resistance genes that encode the aminoglycoside acetyl transferase (AAC) and aminoglycoside adenyl transferase (AAD) enzymes. When the aminoglycosides bind to the leader RNA, a conformational change will be induced, leading to the expression of the aminoglycoside resistance genes ([1] Jia X et al.). In addition, the leader RNA encompass an integrase site (attl1), which is overlapped by a short peptide encoded by an ORF (ORF11) ([2] Roth A et al.). Therefore, Aleader is a complicated riboswitch containing ORF11, cistron, aminoglycoside-binding domain and att1 recombination site. Due to the complicacy of Aleader, the secondary structure of it could not be predicted by most software such as Mfold and NUPACK ([1] Jia X et al.). | Riboswitches are regulatory RNAs that regulate gene expression by binding small molecules. Recently, a novel riboswitch was reported which is present in the leader RNA of the aminoglycoside resistance genes that encode the aminoglycoside acetyl transferase (AAC) and aminoglycoside adenyl transferase (AAD) enzymes. When the aminoglycosides bind to the leader RNA, a conformational change will be induced, leading to the expression of the aminoglycoside resistance genes ([1] Jia X et al.). In addition, the leader RNA encompass an integrase site (attl1), which is overlapped by a short peptide encoded by an ORF (ORF11) ([2] Roth A et al.). Therefore, Aleader is a complicated riboswitch containing ORF11, cistron, aminoglycoside-binding domain and att1 recombination site. Due to the complicacy of Aleader, the secondary structure of it could not be predicted by most software such as Mfold and NUPACK ([1] Jia X et al.). | ||
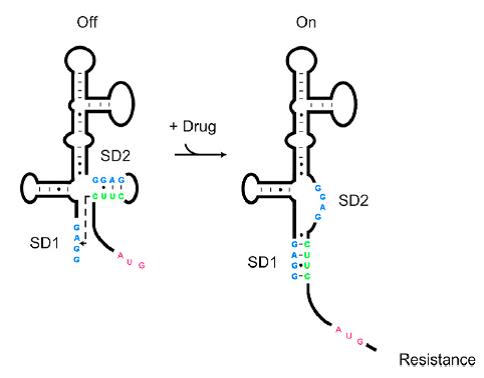
| − | [[File: | + | [[File:AleaderT1.jpg|200px|thumb|left| '''Figure1''' Proposed Model for the Induction of Aminoglycoside Resistance: Aminoglycoside binding to the leader RNA induces a change in the leader RNA structure]] |
As shown in Figure 1, the 75nt RNA sequence of Aleader contains two SD sequences (ribosome binding sites) and an anti-SD sequence (CUUC) which can complementarily pair with either of the SD sequences. In the absence of aminoglycosides, anti-SD pairs with SD2. The binding of ribosomes to SD1 triggers the translation of a small peptide which stops at the stop codon ahead of SD2, and therefore inhibits the translation of the desired gene after SD2. When aminoglycosides (kanamycin for example) exists, it will induce a structural change of Aleader. The anti-SD sequence base-pairs with SD1, consequently unmask SD2 for ribosomal binding, which results in the translation of the following gene. | As shown in Figure 1, the 75nt RNA sequence of Aleader contains two SD sequences (ribosome binding sites) and an anti-SD sequence (CUUC) which can complementarily pair with either of the SD sequences. In the absence of aminoglycosides, anti-SD pairs with SD2. The binding of ribosomes to SD1 triggers the translation of a small peptide which stops at the stop codon ahead of SD2, and therefore inhibits the translation of the desired gene after SD2. When aminoglycosides (kanamycin for example) exists, it will induce a structural change of Aleader. The anti-SD sequence base-pairs with SD1, consequently unmask SD2 for ribosomal binding, which results in the translation of the following gene. | ||
| + | |||
| + | == Our design of AleaderT == | ||
Revision as of 15:14, 26 September 2013
ALeaderT (with insulator)
ALeaderT is a novel riboswitch transformed from the leader RNA of the aminoglycoside resistance genes. It has a low minimal level and a high dynamic range, with a low noise in the system and minimal difference under different regulation.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Background
Riboswitches are regulatory RNAs that regulate gene expression by binding small molecules. Recently, a novel riboswitch was reported which is present in the leader RNA of the aminoglycoside resistance genes that encode the aminoglycoside acetyl transferase (AAC) and aminoglycoside adenyl transferase (AAD) enzymes. When the aminoglycosides bind to the leader RNA, a conformational change will be induced, leading to the expression of the aminoglycoside resistance genes ([1] Jia X et al.). In addition, the leader RNA encompass an integrase site (attl1), which is overlapped by a short peptide encoded by an ORF (ORF11) ([2] Roth A et al.). Therefore, Aleader is a complicated riboswitch containing ORF11, cistron, aminoglycoside-binding domain and att1 recombination site. Due to the complicacy of Aleader, the secondary structure of it could not be predicted by most software such as Mfold and NUPACK ([1] Jia X et al.).
As shown in Figure 1, the 75nt RNA sequence of Aleader contains two SD sequences (ribosome binding sites) and an anti-SD sequence (CUUC) which can complementarily pair with either of the SD sequences. In the absence of aminoglycosides, anti-SD pairs with SD2. The binding of ribosomes to SD1 triggers the translation of a small peptide which stops at the stop codon ahead of SD2, and therefore inhibits the translation of the desired gene after SD2. When aminoglycosides (kanamycin for example) exists, it will induce a structural change of Aleader. The anti-SD sequence base-pairs with SD1, consequently unmask SD2 for ribosomal binding, which results in the translation of the following gene.