Difference between revisions of "Part:BBa K1119009"
| Line 2: | Line 2: | ||
<partinfo>BBa_K1119009 short</partinfo> | <partinfo>BBa_K1119009 short</partinfo> | ||
| − | The CDS of MLS ([[Part:BBa_K1119001|BBa_K1119001]]) was assembled in frame with that of GFP reporter using Freiburg’s RFC25 format([[Part:BBa_K648013|BBa_K648013]]). The translation unit was driven by CMV promoter ([[Part:BBa_K1119006|BBa_K1119006]]) and terminated by hGH polyA signal ([[Part:BBa_K404108|BBa_K404108]]). | + | The CDS of MLS ([[Part:BBa_K1119001|BBa_K1119001]]) was assembled in frame with that of GFP reporter using Freiburg’s RFC25 format([[Part:BBa_K648013|BBa_K648013]]). The translation unit was driven by CMV promoter ([[Part:BBa_K1119006|BBa_K1119006]]) and terminated by hGH polyA signal ([[Part:BBa_K404108|BBa_K404108]]). This construct was used for characterization of MLS BioBrick([[Part:BBa_K1119001|BBa_K1119001]]. |
| + | |||
| + | == Result == | ||
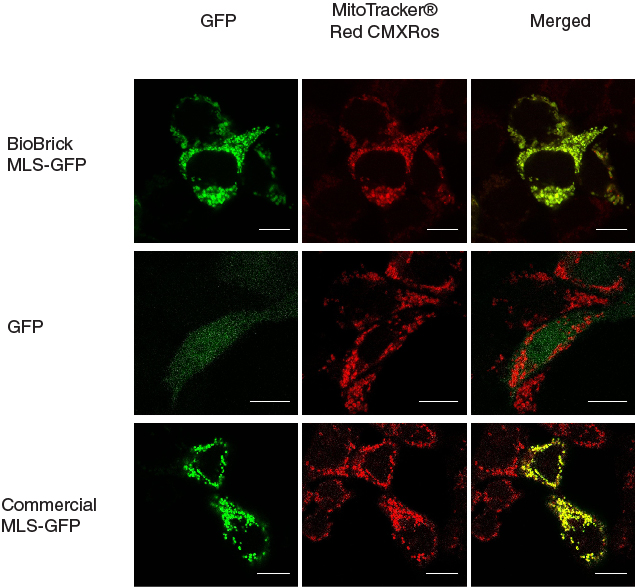
| + | This MLS-GFP generator (BBa_K1119009) was transfected into HEK293FT cells. Mitochondria were stained after transfection and co-localization was determined by area of signal that overlapped. | ||
| + | |||
| + | To provide a positive control, CDS of EGFP from pEGFP-N1 (Clontech) was inserted downstream and in frame with the CDS of the MLS in the commercial plasmid pCMV/myc/mito, (Invitrogen, Carlsbard, CA). A negative control was made by GFP generator that does not contains the CDS of MLS (BBa_K1119008). | ||
[[File:mlschar_1.jpg|600px|thumb|center|'''Figure 1. MLS directs GFP into mitochondria.''' When MLS is added to the N terminus of GFP, the GFP was directed to the mitochondria in the cells, giving patches of GFP signal that overlapped with the signals from MitoTracker®. When MLS is not added to the GFP, the GFP signal can be seen scattered all around in the cell. The contrast for the negative control is up tuned since the raw signal was too dim to be visible. Scale bar = 10 microns]] | [[File:mlschar_1.jpg|600px|thumb|center|'''Figure 1. MLS directs GFP into mitochondria.''' When MLS is added to the N terminus of GFP, the GFP was directed to the mitochondria in the cells, giving patches of GFP signal that overlapped with the signals from MitoTracker®. When MLS is not added to the GFP, the GFP signal can be seen scattered all around in the cell. The contrast for the negative control is up tuned since the raw signal was too dim to be visible. Scale bar = 10 microns]] | ||
Revision as of 09:13, 26 September 2013
CMV promoter - MLS - GFP - hGH polyA tail
The CDS of MLS (BBa_K1119001) was assembled in frame with that of GFP reporter using Freiburg’s RFC25 format(BBa_K648013). The translation unit was driven by CMV promoter (BBa_K1119006) and terminated by hGH polyA signal (BBa_K404108). This construct was used for characterization of MLS BioBrick(BBa_K1119001.
Result
This MLS-GFP generator (BBa_K1119009) was transfected into HEK293FT cells. Mitochondria were stained after transfection and co-localization was determined by area of signal that overlapped.
To provide a positive control, CDS of EGFP from pEGFP-N1 (Clontech) was inserted downstream and in frame with the CDS of the MLS in the commercial plasmid pCMV/myc/mito, (Invitrogen, Carlsbard, CA). A negative control was made by GFP generator that does not contains the CDS of MLS (BBa_K1119008).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 614
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 628
Illegal AgeI site found at 1462 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1865
Illegal BsaI.rc site found at 1367
