Difference between revisions of "Part:BBa K801097"
Nadine1990 (Talk | contribs) (→Background and principles) |
Nadine1990 (Talk | contribs) (→References) |
||
| Line 26: | Line 26: | ||
=References= | =References= | ||
---- | ---- | ||
| − | [http://www.ncbi.nlm.nih.gov/pubmed/ | + | *[[http://www.ncbi.nlm.nih.gov/pubmed/12481418 Gerhauser et al., 2002]] Gerhauser, C., Alt, A., Heiss, E., Gamal-Eldeen, A., Klimo, K., Knauft, J., Neu- mann, I., Scherf, H.-R., Frank, N., Bartsch, H., and Becker, H. (2002). Cancer chemopreventive activity of xanthohumol, a natural product derived from hop. ''Mol Cancer Ther'', 1(11):959–69. |
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/10752639 Henderson et al., 2000]] Henderson, M. C., Miranda, C. L., Stevens, J. F., Deinzer, M. L., and Buhler, D. R. (2000). In vitro inhibition of human p450 enzymes by prenylated flavonoids from hops, ''humulus lupulus''. ''Xenobiotica'', 30(3):235–51. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/14704995 Jiang and Morgan, 2004]] Jiang, H. and Morgan, J. A. (2004). Optimization of an in vivo plant p450 monooxygenase system in ''Saccharomyces cerevisiae''. ''Biotechnol Bioeng'', 85(2):130–7. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/10737704 Miranda et al., 2000a]] Miranda, C. L., Aponso, G. L., Stevens, J. F., Deinzer, M. L., and Buhler, D. R. (2000a). Prenylated chalcones and flavanones as inducers of quinone reductase in mouse hepa 1c1c7 cells. ''Cancer Lett'', 149(1-2):21–9. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/10418944 Miranda et al., 1999]] Miranda, C. L., Stevens, J. F., Helmrich, A., Henderson, M. C., Rodriguez, R. J., Yang, Y. H., Deinzer, M. L., Barnes, D. W., and Buhler, D. R. (1999). Antiproliferative and cytotoxic effects of prenylated flavonoids from hops (''Humulus lupulus'') in human cancer cell lines. ''Food Chem Toxicol'', 37(4):271–85. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/10995285 Miranda et al., 2000b]] Miranda, C. L., Stevens, J. F., Ivanov, V., McCall, M., Frei, B., Deinzer, M. L., and Buhler, D. R. (2000b). Antioxidant and prooxidant actions of prenylated and nonprenylated chalcones and flavanones in vitro. ''J Agric Food Chem'', 48(9):3876–84. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/11038156 Miranda et al., 2000c]] Miranda, C. L., Yang, Y. H., Henderson, M. C., Stevens, J. F., Santana-Rios, G., Deinzer, M. L., and Buhler, D. R. (2000c). Prenylflavonoids from hops inhibit the metabolic activation of the carcinogenic heterocyclic amine 2-amino-3-methylimidazo[4, 5-f]quinoline, mediated by cdna-expressed human cyp1a2. ''Drug Metab Dispos'', 28(11):1297–302. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/19631278 Trantas et al., 2009]] Trantas, E., Panopoulos, N., and Ververidis, F. (2009). Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in ''Saccharomyces cerevisiae''. ''Metab Eng'', 11(6):355–66. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/11240137 Yilmazer et al., 2001a]] Yilmazer, M., Stevens, J. F., and Buhler, D. R. (2001a). In vitro glucuronidation of xanthohumol, a flavonoid in hop and beer, by rat and human liver microsomes. ''FEBS Lett'', 491(3):252–6. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/11181488 Yilmazer et al., 2001b]] Yilmazer, M., Stevens, J. F., Deinzer, M. L., and Buhler, D. R. (2001b). In vitro biotransformation of xanthohumol, a flavonoid from hops (''Humulus lupulus''), by rat liver microsomes. ''Drug Metab Dispos'', 29(3):223–31. | ||
| + | |||
<hr> | <hr> | ||
Revision as of 12:42, 16 October 2012
Chalcone O-methyltransferase (OMT1) + yeast consensus sequence
Chalcone O-methyltransferase (OMT1) coding region from Humulus lupulus preceeded by yeast consensus sequence for improved expression.
Background and principles
Usage and Biology
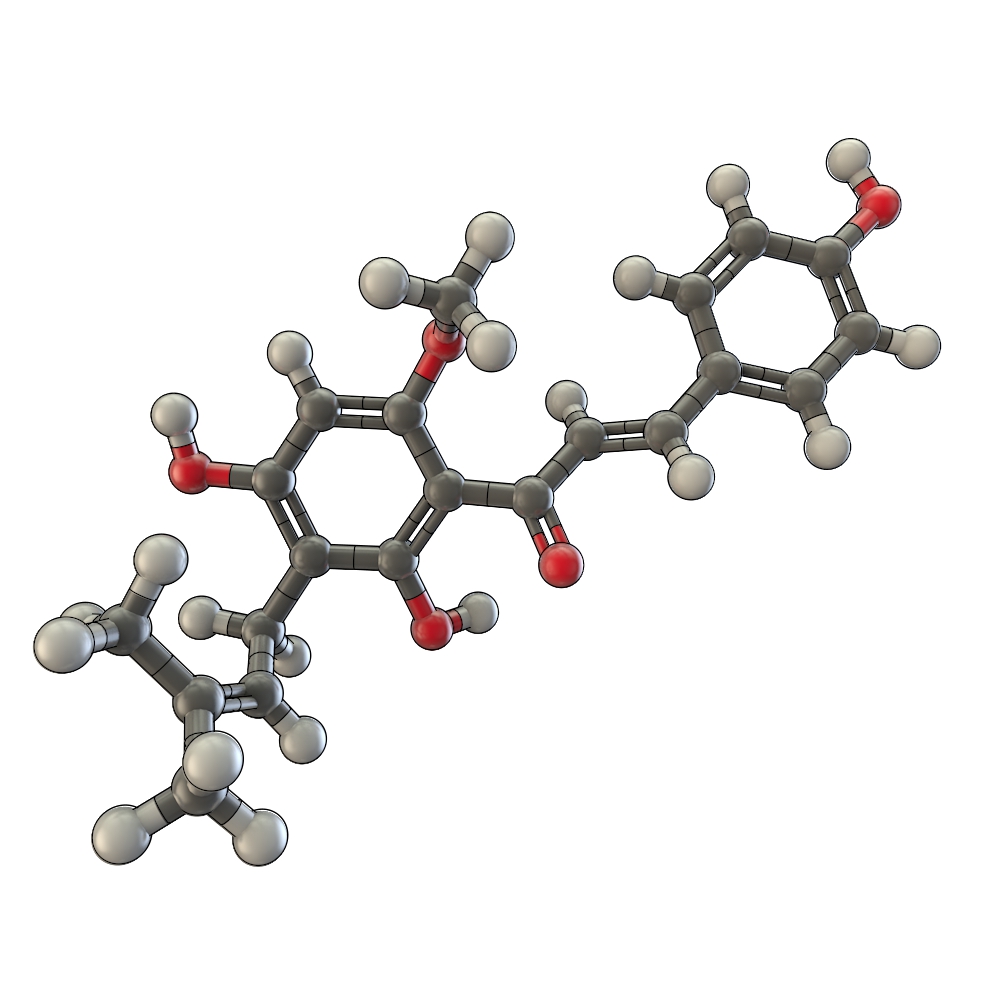
Text on the molecular properties of Xanthohumol.
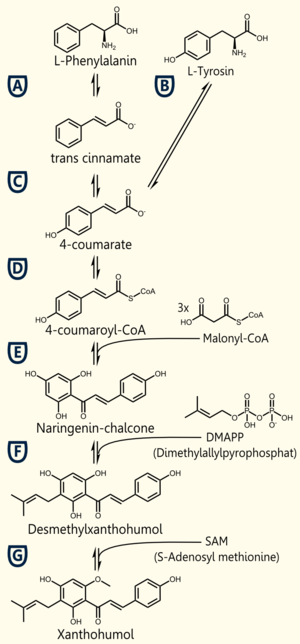
Biosynthesis
Characterization
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 485
Illegal XhoI site found at 300 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
- http://www.ncbi.nlm.nih.gov/pubmed/12481418 Gerhauser et al., 2002 Gerhauser, C., Alt, A., Heiss, E., Gamal-Eldeen, A., Klimo, K., Knauft, J., Neu- mann, I., Scherf, H.-R., Frank, N., Bartsch, H., and Becker, H. (2002). Cancer chemopreventive activity of xanthohumol, a natural product derived from hop. Mol Cancer Ther, 1(11):959–69.
- http://www.ncbi.nlm.nih.gov/pubmed/10752639 Henderson et al., 2000 Henderson, M. C., Miranda, C. L., Stevens, J. F., Deinzer, M. L., and Buhler, D. R. (2000). In vitro inhibition of human p450 enzymes by prenylated flavonoids from hops, humulus lupulus. Xenobiotica, 30(3):235–51.
- http://www.ncbi.nlm.nih.gov/pubmed/14704995 Jiang and Morgan, 2004 Jiang, H. and Morgan, J. A. (2004). Optimization of an in vivo plant p450 monooxygenase system in Saccharomyces cerevisiae. Biotechnol Bioeng, 85(2):130–7.
- http://www.ncbi.nlm.nih.gov/pubmed/10737704 Miranda et al., 2000a Miranda, C. L., Aponso, G. L., Stevens, J. F., Deinzer, M. L., and Buhler, D. R. (2000a). Prenylated chalcones and flavanones as inducers of quinone reductase in mouse hepa 1c1c7 cells. Cancer Lett, 149(1-2):21–9.
- http://www.ncbi.nlm.nih.gov/pubmed/10418944 Miranda et al., 1999 Miranda, C. L., Stevens, J. F., Helmrich, A., Henderson, M. C., Rodriguez, R. J., Yang, Y. H., Deinzer, M. L., Barnes, D. W., and Buhler, D. R. (1999). Antiproliferative and cytotoxic effects of prenylated flavonoids from hops (Humulus lupulus) in human cancer cell lines. Food Chem Toxicol, 37(4):271–85.
- http://www.ncbi.nlm.nih.gov/pubmed/10995285 Miranda et al., 2000b Miranda, C. L., Stevens, J. F., Ivanov, V., McCall, M., Frei, B., Deinzer, M. L., and Buhler, D. R. (2000b). Antioxidant and prooxidant actions of prenylated and nonprenylated chalcones and flavanones in vitro. J Agric Food Chem, 48(9):3876–84.
- http://www.ncbi.nlm.nih.gov/pubmed/11038156 Miranda et al., 2000c Miranda, C. L., Yang, Y. H., Henderson, M. C., Stevens, J. F., Santana-Rios, G., Deinzer, M. L., and Buhler, D. R. (2000c). Prenylflavonoids from hops inhibit the metabolic activation of the carcinogenic heterocyclic amine 2-amino-3-methylimidazo[4, 5-f]quinoline, mediated by cdna-expressed human cyp1a2. Drug Metab Dispos, 28(11):1297–302.
- http://www.ncbi.nlm.nih.gov/pubmed/19631278 Trantas et al., 2009 Trantas, E., Panopoulos, N., and Ververidis, F. (2009). Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metab Eng, 11(6):355–66.
- http://www.ncbi.nlm.nih.gov/pubmed/11240137 Yilmazer et al., 2001a Yilmazer, M., Stevens, J. F., and Buhler, D. R. (2001a). In vitro glucuronidation of xanthohumol, a flavonoid in hop and beer, by rat and human liver microsomes. FEBS Lett, 491(3):252–6.
- http://www.ncbi.nlm.nih.gov/pubmed/11181488 Yilmazer et al., 2001b Yilmazer, M., Stevens, J. F., Deinzer, M. L., and Buhler, D. R. (2001b). In vitro biotransformation of xanthohumol, a flavonoid from hops (Humulus lupulus), by rat liver microsomes. Drug Metab Dispos, 29(3):223–31.