Difference between revisions of "Part:BBa K771001"
| Line 4: | Line 4: | ||
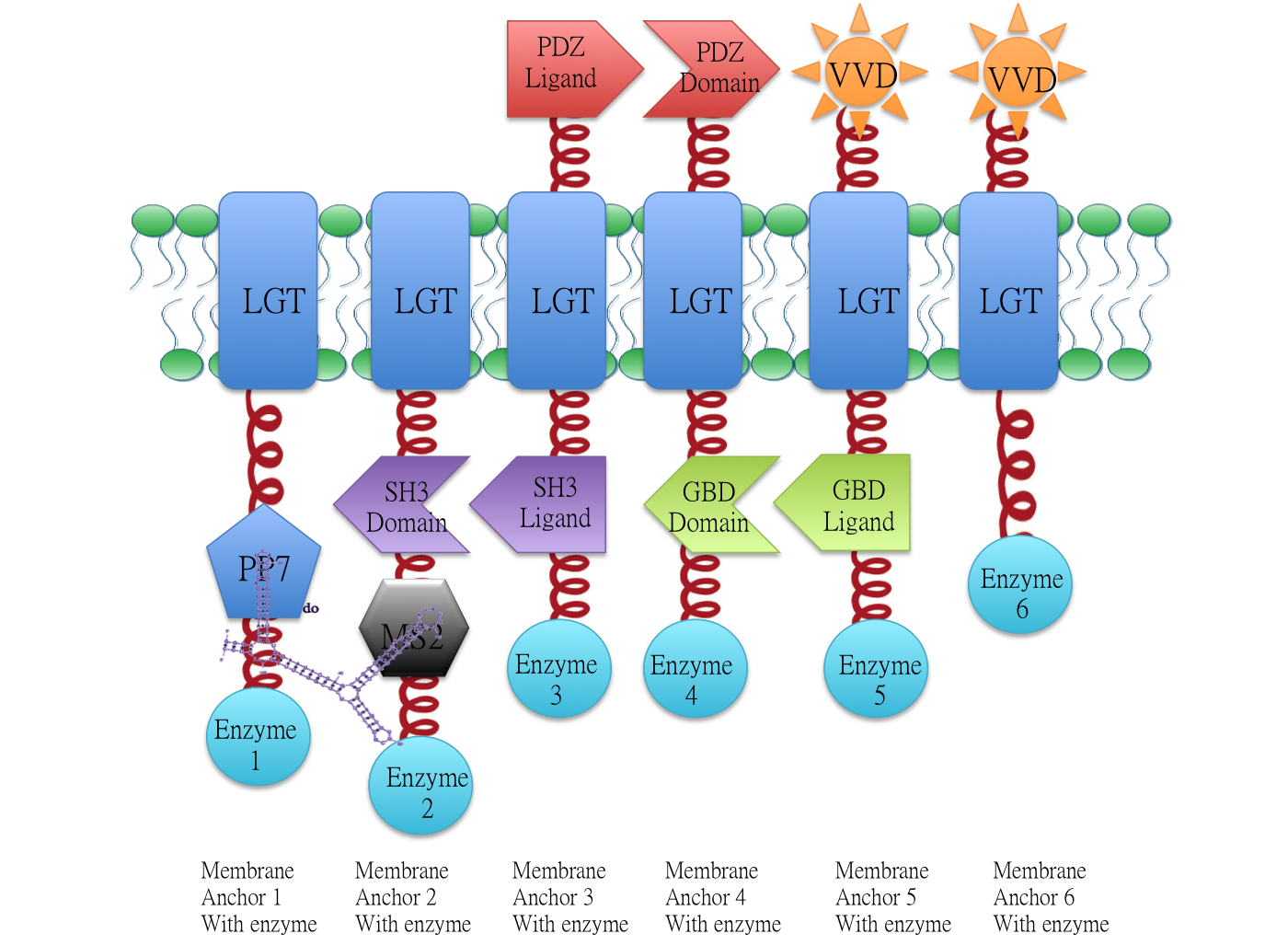
RNA aptamer binding protein PP7 fused with membrane protein Lgt | RNA aptamer binding protein PP7 fused with membrane protein Lgt | ||
[[Image:12SJTU-MA1-MA6.png|500px|center]] | [[Image:12SJTU-MA1-MA6.png|500px|center]] | ||
| + | |||
| + | |||
| + | As there is no compartment in prokaryotic cells, enzymes involved in a biochemical pathway diffuse all over the cytoplasm. Intermediates produced from one enzyme cannot be passed efficiently to the next due to spatial obstacles. | ||
| + | Rationally designed assemblies of enzymes could help substrates flow by decreasing distance that intermediates have to travel between enzymes, and thus increase yields of sequential biological reactions. If we attach those enzymes to engineered membrane proteins which constitutively assemble together, the reactions are going to proceed much faster. | ||
Related Biobrick: | Related Biobrick: | ||
Revision as of 02:46, 28 September 2012
Membrane Anchor 1 :ssDsbA-Lgt-PP7
RNA aptamer binding protein PP7 fused with membrane protein Lgt
As there is no compartment in prokaryotic cells, enzymes involved in a biochemical pathway diffuse all over the cytoplasm. Intermediates produced from one enzyme cannot be passed efficiently to the next due to spatial obstacles.
Rationally designed assemblies of enzymes could help substrates flow by decreasing distance that intermediates have to travel between enzymes, and thus increase yields of sequential biological reactions. If we attach those enzymes to engineered membrane proteins which constitutively assemble together, the reactions are going to proceed much faster.
Related Biobrick: <br\>Membrabe Anchor 2([BBa_K771002]) <br\>Membrabe Anchor 3([BBa_K771003]) <br\>Membrabe Anchor 4([BBa_K771004]) <br\>Membrabe Anchor 5([BBa_K771005]) <br\>Membrabe Anchor 6([BBa_K771006])
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1129
Illegal BglII site found at 1385 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 883
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 504