Difference between revisions of "Part:BBa K777002"
| Line 2: | Line 2: | ||
<partinfo>BBa_K777002 short</partinfo> | <partinfo>BBa_K777002 short</partinfo> | ||
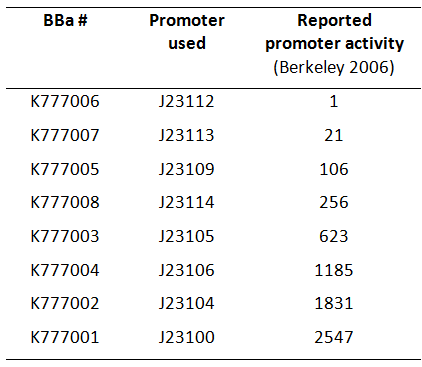
| − | [[Image:Table_K777001-K777008_new.png|thumb| | + | [[Image:Table_K777001-K777008_new.png|thumb|300px|right|'''Fig. 1:''' Overview of BioBricks containing the ''Tar'' gene downstream of Anderson promoters. (Activity according to Berkeley 2006)]] |
The transmembrane chemoreceptor Tar of <i>E. coli</i> mediates chemotaxis towards aspartate and exists as a functional homodimer. | The transmembrane chemoreceptor Tar of <i>E. coli</i> mediates chemotaxis towards aspartate and exists as a functional homodimer. | ||
| − | Here we used a set of 8 constitutive [https://parts.igem.org/Part:BBa_J23104 Anderson promoters] from the 2006 Berkeley group to test how different levels of constitutive ''Tar'' expression affect the chemotaxis of ''E. coli''. | + | Here we used a set of 8 constitutive [https://parts.igem.org/Part:BBa_J23104 Anderson promoters] from the 2006 Berkeley group to test how different levels of constitutive ''Tar'' expression affect the chemotaxis of ''E. coli'' (Fig. 1). |
| − | <br> <br> <br> | + | <br> <br> |
| − | <br> <br> <br> | + | The transcriptional activity of the constructs K777001-K777008 was quantified via quantitative real-time PCR. The results of this experiment can be found in the [https://parts.igem.org/Part:BBa_J23100:Experience#User_Reviews experience section] of the according Anderson promoters. Our data were compared to the activity of the promoters that can be found on the main page of [https://parts.igem.org/Part:BBa_J23100 Anderson promoters].<br> The respective raw dataset can be found in our Team-wiki [http://2012.igem.org/Team:Goettingen/Project/Computational_Data#Raw_Datasets here]. |
| − | <br> <br> | + | <br> <br><br> <br> <br><br> <br> <br> |
| − | <br> <br> <br> | + | |
| Line 16: | Line 15: | ||
* Here is the complete sequence of this part as an [https://static.igem.org/mediawiki/2012/f/fc/Tar_receptor_under_the_control_of_constitutive_promoter.zip ApE file]. | * Here is the complete sequence of this part as an [https://static.igem.org/mediawiki/2012/f/fc/Tar_receptor_under_the_control_of_constitutive_promoter.zip ApE file]. | ||
<br> | <br> | ||
| + | ====Chemotaxis functionality==== | ||
| + | [[Image:Gruppe3_Figure02neu.png|500px|thumb|right|'''Fig. 2:''' Promoter comparison through a chemotaxis assay.]] | ||
| + | The different promoter constructs K777001-K777008 were all tested for chemotaxis towards aspartate (Fig. 2). | ||
| + | Eight promoters of the „Anderson“ collection were cloned in front of the gene coding for the Tar receptor inside the pSB1C3 backbone. Here, we compared the swimming behavior of these strains in chemotaxis assay with M9-swimming agar plates. The chemotaxis assay was conducted as described in the methods section. L-aspartate served as the attractant in the central Whatman paper. The numbers next to each halo correspond to the promoter activities reported by the Berkley University in 2006. <b>(A)</b> The constructs were transformed in the Δtar strain. The picture was taken after 3 days incubation at 33 °C. The halos of the strongest promoters (J23100, x2547 and J23104, x1831) had a slightly bigger radius than the other promoter constructs. In addition, slight chemotaxis could be observed for these strains. The drop of the construct J23106 was smeared during the application and is therefore not evaluable. <b>(B)</b> The constructs were transformed in the BL21 strain, that naturally expresses the Tar receptor. The picture was taken after 1 day incubation at 33 °C. In general, halo size did not vary strongly. The halos of the strongest promoters (J23100, x2547 and J23104, x1831) had the biggest radius. The halo of J23106 (x1185) showed the smallest radius, although this promoter has the third stronges activity. Chemotaxis could be observed for every strain. | ||
| + | <br><br><br> | ||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K777002 SequenceAndFeatures</partinfo> | <partinfo>BBa_K777002 SequenceAndFeatures</partinfo> | ||
<br> | <br> | ||
| − | [[Image:K777002_in_pSB1C3.png|frame|left|'''Fig. | + | [[Image:K777002_in_pSB1C3.png|frame|left|'''Fig. 3:''' Plasmid map of K777002 in pSB1C3]] |
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Revision as of 20:15, 26 September 2012
Tar receptor under the control of constitutive promoter J23104
The transmembrane chemoreceptor Tar of E. coli mediates chemotaxis towards aspartate and exists as a functional homodimer.
Here we used a set of 8 constitutive Anderson promoters from the 2006 Berkeley group to test how different levels of constitutive Tar expression affect the chemotaxis of E. coli (Fig. 1).
The transcriptional activity of the constructs K777001-K777008 was quantified via quantitative real-time PCR. The results of this experiment can be found in the experience section of the according Anderson promoters. Our data were compared to the activity of the promoters that can be found on the main page of Anderson promoters.
The respective raw dataset can be found in our Team-wiki [http://2012.igem.org/Team:Goettingen/Project/Computational_Data#Raw_Datasets here].
Usage and Biology
This part can be used for chemotaxis assays.
- Here is the complete sequence of this part as an ApE file.
Chemotaxis functionality
The different promoter constructs K777001-K777008 were all tested for chemotaxis towards aspartate (Fig. 2).
Eight promoters of the „Anderson“ collection were cloned in front of the gene coding for the Tar receptor inside the pSB1C3 backbone. Here, we compared the swimming behavior of these strains in chemotaxis assay with M9-swimming agar plates. The chemotaxis assay was conducted as described in the methods section. L-aspartate served as the attractant in the central Whatman paper. The numbers next to each halo correspond to the promoter activities reported by the Berkley University in 2006. (A) The constructs were transformed in the Δtar strain. The picture was taken after 3 days incubation at 33 °C. The halos of the strongest promoters (J23100, x2547 and J23104, x1831) had a slightly bigger radius than the other promoter constructs. In addition, slight chemotaxis could be observed for these strains. The drop of the construct J23106 was smeared during the application and is therefore not evaluable. (B) The constructs were transformed in the BL21 strain, that naturally expresses the Tar receptor. The picture was taken after 1 day incubation at 33 °C. In general, halo size did not vary strongly. The halos of the strongest promoters (J23100, x2547 and J23104, x1831) had the biggest radius. The halo of J23106 (x1185) showed the smallest radius, although this promoter has the third stronges activity. Chemotaxis could be observed for every strain.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 1323
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 152