Difference between revisions of "Part:BBa K801020"
Simon Heinze (Talk | contribs) (→Usage and Biology) |
Simon Heinze (Talk | contribs) (→Usage and Biology) |
||
| Line 20: | Line 20: | ||
[[Image:TUM12_KlADH4_eGFP-expression_Graph1.jpg|600px]] | [[Image:TUM12_KlADH4_eGFP-expression_Graph1.jpg|600px]] | ||
| + | |||
| + | The result | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
Revision as of 15:18, 24 September 2012
KlADH4 yeast promoter, ethanol inducible
This part is the ethanol inducible promoter controlling the KlADH4-gene of K. lactis.
The use of this ethanol inducible promoter to produce heterologous proteins in K. lactis was shown by Salioa et al. 1999 [http://www.ncbi.nlm.nih.gov/pubmed?term=9872759].
We characterized this part in S. cerevisiae (strain INVSc1) to find out whether this part is also ethanol-inducible in this yeast.
Usage and Biology
The UASe-region of this promoter has been shown to be responsible for the ethanol sensitivity of this promoter in K. lactis (Mazzoni et al., 2000 [http://www.ncbi.nlm.nih.gov/pubmed?term=10724480]). The region includes binding sites for the Rap1-protein (repressor activator protein 1) and the Yap1-protein (a transcription factor involved in stress response) as well as two heat shock elements (HSE) and five stress response elemtents (STRE). All these cis-elements and the respecitve proteins also occur in S. cerevisiae. For this reason we wanted to examine whether the KlADH4-promoter remains ethanol inducible if it is transferred from its natural organism (K. lactis) to S. cerevisiae.
The characterization of this part was done using a KlADH4-promoter + eGFP construct.
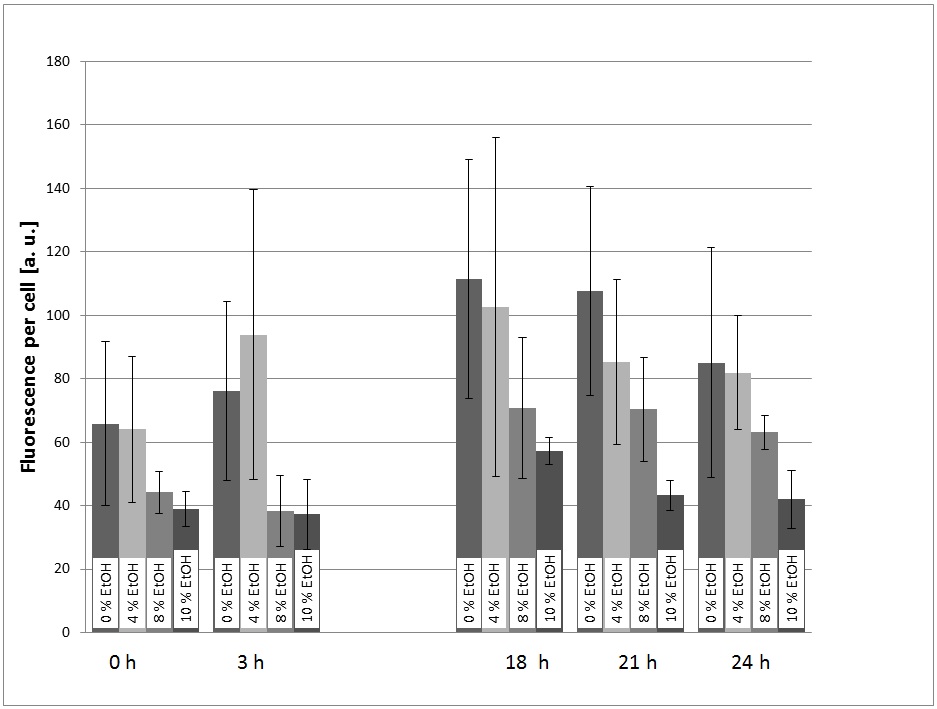
In a first experiment, the transformed yeast cells were picked grown in a pre-culture (SC-U Medium) over night and transferred into SC-U Medium with different concentrations of ethanol (0%, 4%, 8%, 10%). The eGFP-fluorescence and the OD600 were measured at t = 0h, 3h, 18h, 21h, 24h.
For the evaluation of the experimental data, the measured fluorescence was divided by the respecitve OD600, to normalize the fluorescence to the respecitve cell count. This was done to take the intrinsic auto-fluorescence in account. The results are shown below:
The result
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 753
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 590
References
- [1] Saliola, M., Mazzoni, C., Solimando, N., Crisà, A., Falcone, C. & Jung, G. (1999) ‚Use of the KlADH4 promoter for ethanol-dependent production of recombinant human serum albumin in Kluyveromyces lactis’, Appl Environ Microbiol. 65 (1), 53-60. [http://www.ncbi.nlm.nih.gov/pubmed?term=10724480 PMID: 9872759]
- [2] Mazzoni, C., Santori, F., Saliola, M. & Falcone, C. (2000) ‚Molecular analysis of UASE, a cis element containing stress response elements responsible for ethanol induction of the KlADH4 gene of Kluyveromyces lactis’, Res. Microbiol. 151, 19-28. [http://www.ncbi.nlm.nih.gov/pubmed?term=10724480 PMID: 10724480]