Difference between revisions of "Part:BBa K510022:Experience"
(→Characterization of the "portable" attTn7) |
(→Characterization of the "portable" attTn7) |
||
| Line 5: | Line 5: | ||
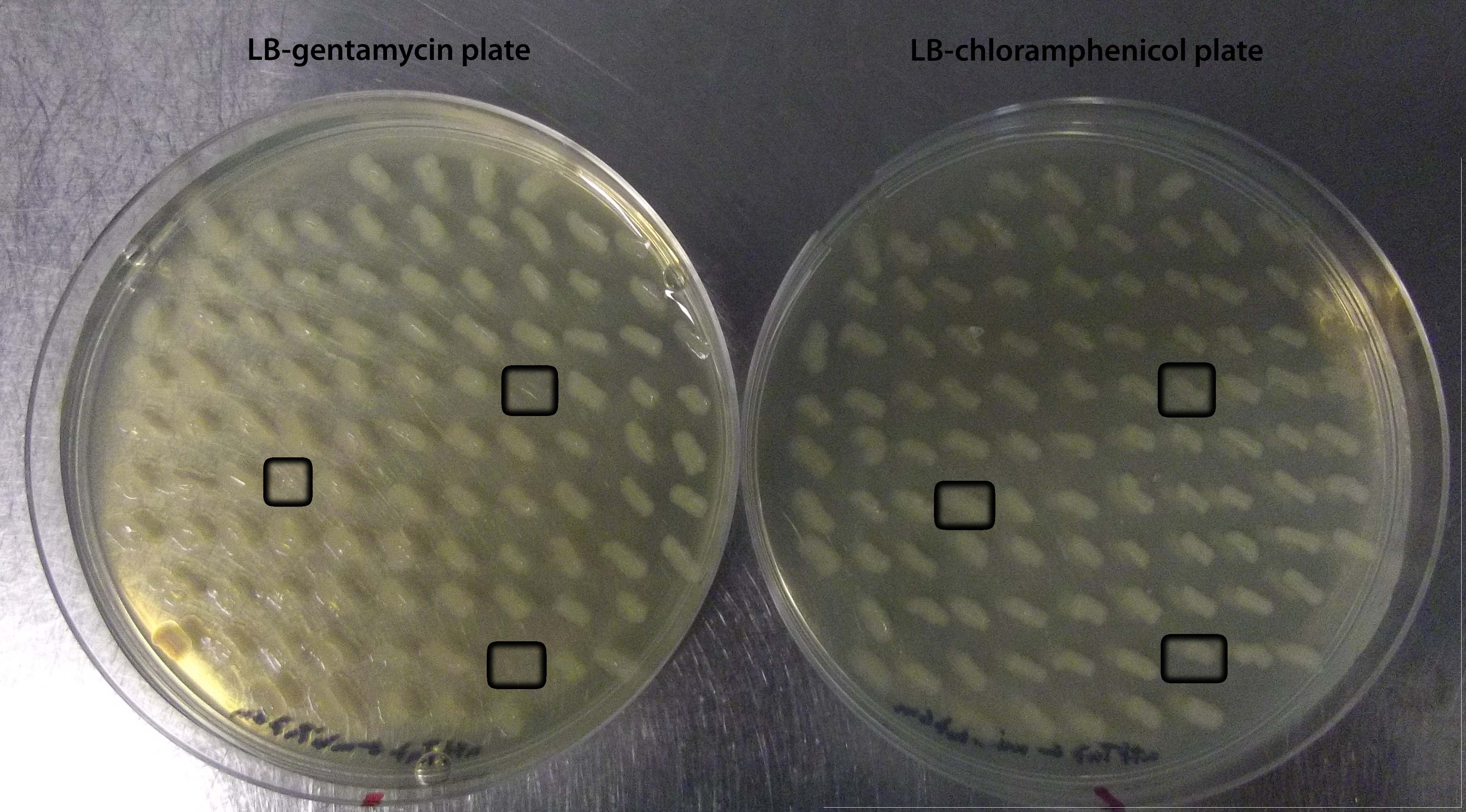
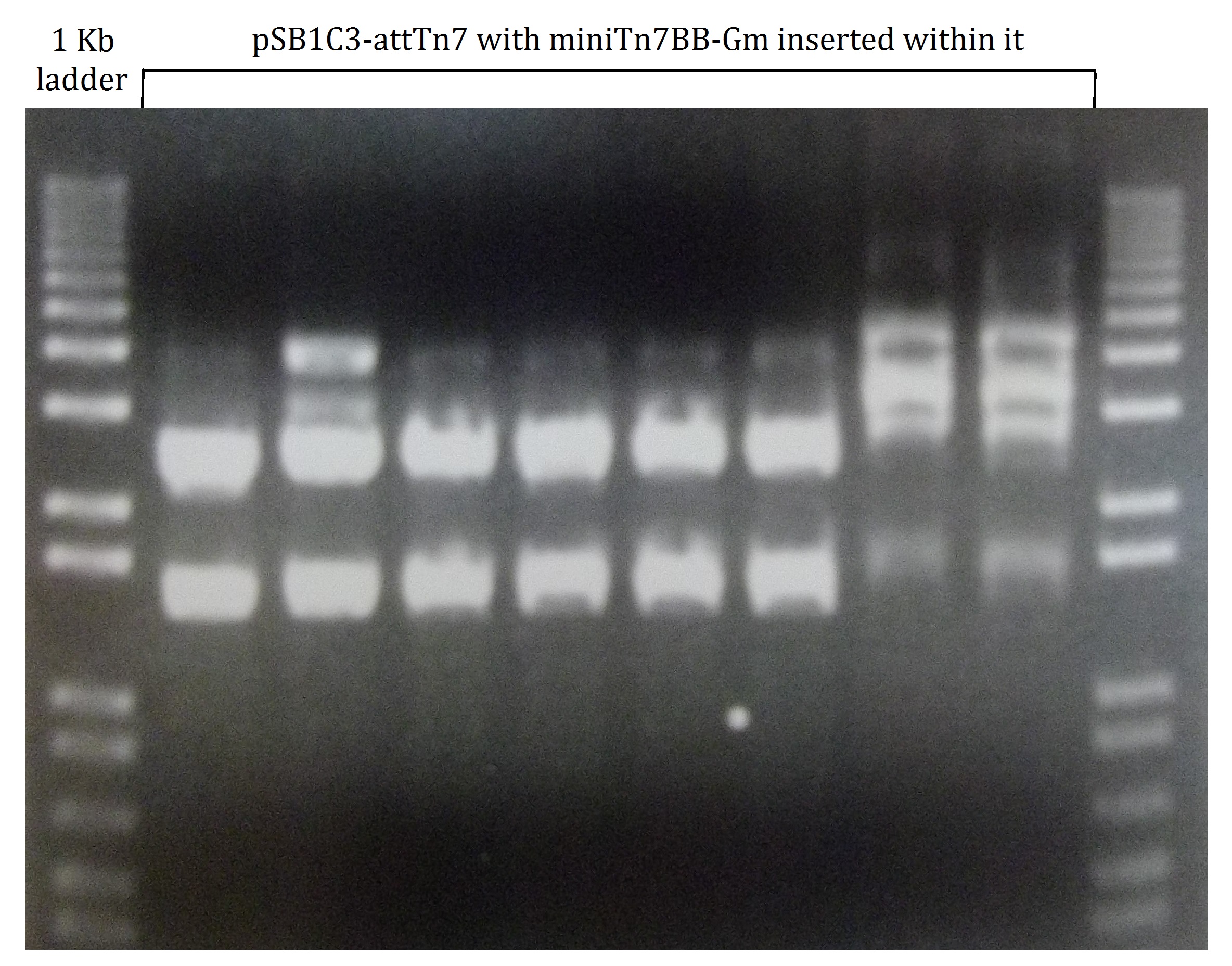
For preliminary characterization of the "portable" attTn7, we attempted to demonstrate transposition into the plasmid-borne target. The miniTn7BB-Gm transposon was transferred into E. coli DH5α bearing pSB1C3-attTn7 by '''tetraparental mating''' using the helper plasmid pTNS2 to provide the Tn7 transposition functions. Transconjugants bearing transposon insertions were selected on LB plates supplemented with gentamycin and chloramphenicol. To identify clones bearing the transposon insertion at the plasmid-borne target, transconjugant colonies were pooled, plasmid DNA was extracted from the pool and used to transform E. coli. Transformants were selected on LB plates containing chloramphenicol and scored for gentamycin resistance (Figure 1). '''97 out of 100''' colonies had become simultaneously resistant to both antibiotics, strongly suggesting that the miniTn7BB-Gm was inserted in the chloramphenicol resistance plasmid, and transferred along with its target. Restriction analysis confirmed that indeed this was the case, and that the transposon was inserted within the attTn7 target site at the pSB1C3-attTn7 plasmid (Figure 2). | For preliminary characterization of the "portable" attTn7, we attempted to demonstrate transposition into the plasmid-borne target. The miniTn7BB-Gm transposon was transferred into E. coli DH5α bearing pSB1C3-attTn7 by '''tetraparental mating''' using the helper plasmid pTNS2 to provide the Tn7 transposition functions. Transconjugants bearing transposon insertions were selected on LB plates supplemented with gentamycin and chloramphenicol. To identify clones bearing the transposon insertion at the plasmid-borne target, transconjugant colonies were pooled, plasmid DNA was extracted from the pool and used to transform E. coli. Transformants were selected on LB plates containing chloramphenicol and scored for gentamycin resistance (Figure 1). '''97 out of 100''' colonies had become simultaneously resistant to both antibiotics, strongly suggesting that the miniTn7BB-Gm was inserted in the chloramphenicol resistance plasmid, and transferred along with its target. Restriction analysis confirmed that indeed this was the case, and that the transposon was inserted within the attTn7 target site at the pSB1C3-attTn7 plasmid (Figure 2). | ||
| − | [[Image: | + | [[Image:UPOSevilla-attTn7-miniTn7-plates.png|600px|center]] |
'''Figure 1. Patches of E. coli transformed with plasmid DNA from pooled colonies obtained after insertion of pUC18Sfi-miniTn7BB-Gm into the genome of ''E.coli'' DH5α bearing pSC1C3-attTn7'''. Open squares indicate patches showing sensitivity to gentamycin, that is, '''not bearing''' the miniTn7 transposon insertion. | '''Figure 1. Patches of E. coli transformed with plasmid DNA from pooled colonies obtained after insertion of pUC18Sfi-miniTn7BB-Gm into the genome of ''E.coli'' DH5α bearing pSC1C3-attTn7'''. Open squares indicate patches showing sensitivity to gentamycin, that is, '''not bearing''' the miniTn7 transposon insertion. | ||
| − | [[Image: | + | [[Image:UPOSevilla-attTn7-miniTn7-gel.png|600px|center]] |
'''Figure 2. Agarose gel of restriction analysis products of eight different samples of pSB1C3-attTn7 with miniTn7BB-Gm module inserted within it'''. Whether the insertion occurs site-specifically in the insertion site of the portable attTn7 is checked by the presence of 2497bp and 1485bp DNA fragments. These restriction products can be easily find in the first six digestion products. The next two looks to be right too, but impurities in their minipreps may make the digestions difficult to understand. We can also see a partial digestion in the second lane. | '''Figure 2. Agarose gel of restriction analysis products of eight different samples of pSB1C3-attTn7 with miniTn7BB-Gm module inserted within it'''. Whether the insertion occurs site-specifically in the insertion site of the portable attTn7 is checked by the presence of 2497bp and 1485bp DNA fragments. These restriction products can be easily find in the first six digestion products. The next two looks to be right too, but impurities in their minipreps may make the digestions difficult to understand. We can also see a partial digestion in the second lane. | ||
Revision as of 18:20, 29 October 2011
Contents
Construction of a "portable" attTn7
As a first step toward this goal, we undertook the cloning of the functional E. coli attTn7 into a plasmid vector, and the demonstration that this "portable" plasmid-borne attTn7 can be recognized as a Tn7 transposition target by our miniTn7BB minitransposons. The attTn7 was PCR-amplified from the chromosome of the E. coli K12 strain MC4100 with primers bearing prefix and suffix restriction sites, cleaved and ligated into pSB1C3. The construct was verified by sequencing.
Characterization of the "portable" attTn7
For preliminary characterization of the "portable" attTn7, we attempted to demonstrate transposition into the plasmid-borne target. The miniTn7BB-Gm transposon was transferred into E. coli DH5α bearing pSB1C3-attTn7 by tetraparental mating using the helper plasmid pTNS2 to provide the Tn7 transposition functions. Transconjugants bearing transposon insertions were selected on LB plates supplemented with gentamycin and chloramphenicol. To identify clones bearing the transposon insertion at the plasmid-borne target, transconjugant colonies were pooled, plasmid DNA was extracted from the pool and used to transform E. coli. Transformants were selected on LB plates containing chloramphenicol and scored for gentamycin resistance (Figure 1). 97 out of 100 colonies had become simultaneously resistant to both antibiotics, strongly suggesting that the miniTn7BB-Gm was inserted in the chloramphenicol resistance plasmid, and transferred along with its target. Restriction analysis confirmed that indeed this was the case, and that the transposon was inserted within the attTn7 target site at the pSB1C3-attTn7 plasmid (Figure 2).
Figure 1. Patches of E. coli transformed with plasmid DNA from pooled colonies obtained after insertion of pUC18Sfi-miniTn7BB-Gm into the genome of E.coli DH5α bearing pSC1C3-attTn7. Open squares indicate patches showing sensitivity to gentamycin, that is, not bearing the miniTn7 transposon insertion.
Figure 2. Agarose gel of restriction analysis products of eight different samples of pSB1C3-attTn7 with miniTn7BB-Gm module inserted within it. Whether the insertion occurs site-specifically in the insertion site of the portable attTn7 is checked by the presence of 2497bp and 1485bp DNA fragments. These restriction products can be easily find in the first six digestion products. The next two looks to be right too, but impurities in their minipreps may make the digestions difficult to understand. We can also see a partial digestion in the second lane.
Applications of BBa_K510022
The possibility of inserting a functional attTn7 in the genomes of organisms without a suitable Tn7 target may enable site-specific Tn7 transposition for at least some of them, thus expanding the host range of this transposon and its derived tools, such as the miniTn7 BioBrick toolkit.
User Reviews
UNIQ6a34182f2ebcf3dc-partinfo-00000000-QINU UNIQ6a34182f2ebcf3dc-partinfo-00000001-QINU