Difference between revisions of "Part:BBa K640002:Experience"
Emily Hicks (Talk | contribs) |
|||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | ||
| Line 5: | Line 4: | ||
===Applications of BBa_K640002=== | ===Applications of BBa_K640002=== | ||
| + | |||
| + | <html> | ||
| + | |||
| + | <h2>Characterization</h2> | ||
| + | |||
| + | <h3>Functionality Test</h3> | ||
| + | |||
| + | In our first assay, we wanted to see that our BBa_K640004 (oriT-ori1600) construct was working. We incubated the co-cutures for 12 hours at 37°c prior to plating. The plates were then left to grow overnight. | ||
| + | |||
| + | <h3>Results</h3> | ||
| + | |||
| + | </html>[[image:Ucalgary_2011_MacConkiePlates.png]]<html> | ||
| + | <br><br> | ||
| + | The large plates on the right show controls of <i>Pseudomonas</i> and <i>E. coli</i> on MacConkey plates indicating the normal expected colors for these organisms. The small plates on the bottom represent our construct, conjugated with two different strains of pseudomonas and plated on MacConkey agar with Kanamycin (50ug/mL). The yellow color indicates that the colonies are <i>Pseudomonas</i>. The small plates on the top show our two strains of <i>Pseudomonas</i>, as well as <i>E. coli</i> without kanamycin resistance plated on MacConkey agar plates with kanamycin (50 ug/mL). All of these plates are pink in color, and show no colonies. This was expected, as it shows that without a resistance marker, these organisms are not able to grow on the MacConkey kanamycin plates. This demonstrates that our part is working the way we expect it to! | ||
| + | <br> | ||
| + | <br> | ||
| + | <h3>Time Assay</h3> | ||
| + | |||
| + | Once we had demonstrated that our part was working, we went on to try to determine how long it would take for the conjugation to occur. We experimented with different incubation times of the co-cultures at 37° before plating. We tried incubating after 10 minutes, 2 hours, 6 hours and 12 hours. | ||
| + | <br> | ||
| + | <h3>Results</h3> | ||
| + | |||
| + | The table below shows the colonie counts that we recorded after each interval. Counts are the average of two repeats. | ||
| + | |||
| + | <table border="1"> | ||
| + | |||
| + | <tr> | ||
| + | <td style="padding-left: 10px; padding-right: 10px;"><p><b>Incubation<br>Time</b></p></td> | ||
| + | <td style="padding-left: 10px; padding-right: 10px;"><p><b>Average #<br>of Colonies</b></p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="padding-left: 10px; padding-right: 10px;"><p>10 min</p></td> | ||
| + | <td style="padding-left: 10px; padding-right: 10px;"><p>0</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="padding-left: 10px; padding-right: 10px;"><p>2 hrs</p></td> | ||
| + | <td style="padding-left: 10px; padding-right: 10px;"><p>2</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="padding-left: 10px; padding-right: 10px;"><p>6 hrs</p></td> | ||
| + | <td style="padding-left: 10px; padding-right: 10px;"><p>4.5</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="padding-left: 10px; padding-right: 10px;"><p>12 hrs</p></td> | ||
| + | <td style="padding-left: 10px; padding-right: 10px;"><p>150</p></td> | ||
| + | </tr> | ||
| + | |||
| + | </table> | ||
| + | |||
| + | |||
| + | </table> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | <h3>Comparison between BBa_K640004 and BBa_K640005</h3> | ||
| + | |||
| + | As we mentioned before, there is an existing oriT in the registry (BBa_J01003), that the 2008 Heidelberg team had previously characterized in 2008 for the conjugation of plasmids in <i>E. coli</i>. This is the OriT that is present in our BBa_K640006 construct, which contains the oriT and our ori1600 (BBa_K640003). We had hypothesized that our OriT, given its longer sequence, may contain elements that improve the efficiency of conjugation to <i>Pseudomonas spp</i>. To test this, we did an assay using <i>E. coli</i> conatining our BBa_K640004 part (which contains our oriT: BBa_K640002 and BBa_K640003) as well as <i>E. coli</i> containing BBa_K640006. | ||
| + | |||
| + | We used a simpler conjugation assay, adapted from <i>Serna et al.</i>, 2010. Overnight cultures of the donor and the recipient were prepared in LB medium. Aliquots of 500 μl were mixed to obtain a donor/recipient ratio of 1:1. Each mixture was centrifuged to pellet the cells, and the supernatant was discarded. The cells were resuspended in 50 μl of LB broth. Mating mixtures were incubated at 37° for 4 h. Diluted and undiluted aliquots were then spread on selective plates. | ||
| + | |||
| + | <h3>Results</h3> | ||
| + | <br> | ||
| + | |||
| + | </html>[[Image:UCALGARY_2011_IMG_0535.jpg|400px]]<html> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | The image below shows our results. On the top, <i>E. coli</i> cells containing the BBa_K640004 construct (which contains our oriT) were used. On the bottom row, on either end, we see plates where <i>E. coli</i> cells containing a kanamycin resistant plasmid, but no OriT or ori1600 construct were used. These are pink in color, with no cells growing,indicating that <i>Pseudomonas</i> and <i>E. coli</i> are not able to naturally conjugate plasmids without our part. In the middle of the bottom row, we see plates where <i>E. coli</i> cells containing the BBa_K640005 construct was used (where we use the registry oriT with our ori1600). As we can see, the plates with the BBa_K640004 are much lighter in color, than the plates where the BBa_K640005 construct was used. This is indicative that there is increased pseudomonas growth, when we use our oriT, as compared to the registry oriT. This was exactly what we had expected, and we hope to have more quantitative data in the next few weeks! | ||
| + | |||
| + | </html> | ||
===User Reviews=== | ===User Reviews=== | ||
Revision as of 03:44, 29 September 2011
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K640002
Characterization
Functionality Test
In our first assay, we wanted to see that our BBa_K640004 (oriT-ori1600) construct was working. We incubated the co-cutures for 12 hours at 37°c prior to plating. The plates were then left to grow overnight.Results
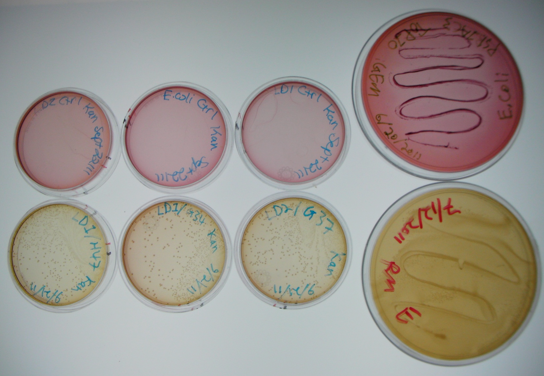
The large plates on the right show controls of Pseudomonas and E. coli on MacConkey plates indicating the normal expected colors for these organisms. The small plates on the bottom represent our construct, conjugated with two different strains of pseudomonas and plated on MacConkey agar with Kanamycin (50ug/mL). The yellow color indicates that the colonies are Pseudomonas. The small plates on the top show our two strains of Pseudomonas, as well as E. coli without kanamycin resistance plated on MacConkey agar plates with kanamycin (50 ug/mL). All of these plates are pink in color, and show no colonies. This was expected, as it shows that without a resistance marker, these organisms are not able to grow on the MacConkey kanamycin plates. This demonstrates that our part is working the way we expect it to!
Time Assay
Once we had demonstrated that our part was working, we went on to try to determine how long it would take for the conjugation to occur. We experimented with different incubation times of the co-cultures at 37° before plating. We tried incubating after 10 minutes, 2 hours, 6 hours and 12 hours.Results
The table below shows the colonie counts that we recorded after each interval. Counts are the average of two repeats.Incubation |
Average # |
10 min |
0 |
2 hrs |
2 |
6 hrs |
4.5 |
12 hrs |
150 |
Comparison between BBa_K640004 and BBa_K640005
As we mentioned before, there is an existing oriT in the registry (BBa_J01003), that the 2008 Heidelberg team had previously characterized in 2008 for the conjugation of plasmids in E. coli. This is the OriT that is present in our BBa_K640006 construct, which contains the oriT and our ori1600 (BBa_K640003). We had hypothesized that our OriT, given its longer sequence, may contain elements that improve the efficiency of conjugation to Pseudomonas spp. To test this, we did an assay using E. coli conatining our BBa_K640004 part (which contains our oriT: BBa_K640002 and BBa_K640003) as well as E. coli containing BBa_K640006. We used a simpler conjugation assay, adapted from Serna et al., 2010. Overnight cultures of the donor and the recipient were prepared in LB medium. Aliquots of 500 μl were mixed to obtain a donor/recipient ratio of 1:1. Each mixture was centrifuged to pellet the cells, and the supernatant was discarded. The cells were resuspended in 50 μl of LB broth. Mating mixtures were incubated at 37° for 4 h. Diluted and undiluted aliquots were then spread on selective plates.Results

The image below shows our results. On the top, E. coli cells containing the BBa_K640004 construct (which contains our oriT) were used. On the bottom row, on either end, we see plates where E. coli cells containing a kanamycin resistant plasmid, but no OriT or ori1600 construct were used. These are pink in color, with no cells growing,indicating that Pseudomonas and E. coli are not able to naturally conjugate plasmids without our part. In the middle of the bottom row, we see plates where E. coli cells containing the BBa_K640005 construct was used (where we use the registry oriT with our ori1600). As we can see, the plates with the BBa_K640004 are much lighter in color, than the plates where the BBa_K640005 construct was used. This is indicative that there is increased pseudomonas growth, when we use our oriT, as compared to the registry oriT. This was exactly what we had expected, and we hope to have more quantitative data in the next few weeks!
User Reviews
UNIQ55c46f8570de78ab-partinfo-00000003-QINU UNIQ55c46f8570de78ab-partinfo-00000004-QINU
