Difference between revisions of "Part:BBa C0060:Experience"
(→User Reviews) |
(→User Reviews) |
||
| Line 6: | Line 6: | ||
===User Reviews=== | ===User Reviews=== | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- DON'T DELETE --><partinfo>BBa_C0060 StartReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_C0060 StartReviews</partinfo> | ||
<!-- Template for a user review | <!-- Template for a user review | ||
| Line 26: | Line 12: | ||
|width='10%'| | |width='10%'| | ||
<partinfo>BBa_C0060 AddReview 1</partinfo> | <partinfo>BBa_C0060 AddReview 1</partinfo> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<I>Username</I> | <I>Username</I> | ||
|width='60%' valign='top'| | |width='60%' valign='top'| | ||
| Line 81: | Line 42: | ||
<br> | <br> | ||
| − | A system of differential equations ( | + | A system of differential equations has been derived and we tried to identify the parameters, studying the exponential growth phase: |
| + | |||
| + | <div style='text-align:justify'> | ||
| + | <div class="thumbinner" style="width: 850px;"> | ||
| + | <a href="https://static.igem.org/mediawiki/2011/1/11/Pc_aiia.jpg"> | ||
| + | <img alt="" src="https://static.igem.org/mediawiki/2011/1/11/Pc_aiia.jpg" class="thumbimage" height="70%" width="68%"></a> | ||
| + | </div></div> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | |||
| + | The four measurment parts ptet-RBSx-LuxI were quantitatively characetrized using the <a href="https://parts.igem.org/Part:BBa_T9002:Experience#t9002">BBa_T9002 biosensor</a>. Unfortunately we were not able to estimate the model parameters (k<sub>cat</sub>, k<sub>M,AiiA</sub>) because placing the device in low copy plasmid (<a href="https://parts.igem.org/Part:pSB4C5">pSB4C5</a>) we did not observe HSL degradation, as you can see in the figures below: | ||
| + | |||
| + | <table width='100%'><tr><td width='50%'> | ||
| + | <div style="text-align:justify"><div class="thumbinner" width='100%'> | ||
| + | <a href="https://static.igem.org/mediawiki/2011/6/69/Aiia_LC_B0030.jpg"> | ||
| + | <img alt="" src="https://static.igem.org/mediawiki/2011/6/69/Aiia_LC_B0030.jpg" class="thumbimage" width="100%"></a></div></div> | ||
| + | </td> | ||
| + | <td width='50%'> | ||
| + | <div style="text-align:justify"><div class="thumbinner" width='100%'> | ||
| + | <a href="https://static.igem.org/mediawiki/2011/6/63/Aiia_LC_B0031.jpg"> | ||
| + | <img alt="" src="https://static.igem.org/mediawiki/2011/6/63/Aiia_LC_B0031.jpg" class="thumbimage" width="100%"></a></div></div> | ||
| + | </td></tr> | ||
| + | <tr><td width='50%'> | ||
| + | <div style="text-align:justify"><div class="thumbinner" width='100%'> | ||
| + | <a href="https://static.igem.org/mediawiki/2011/f/f5/Aiia_LC_B0032.jpg"> | ||
| + | <img alt="" src="https://static.igem.org/mediawiki/2011/f/f5/Aiia_LC_B0032.jpg" class="thumbimage" width="100%"></a></div></div> | ||
| + | </td> | ||
| + | <td width='50%'> | ||
| + | <div style="text-align:justify"><div class="thumbinner" width='100%'> | ||
| + | <a href="https://static.igem.org/mediawiki/2011/8/8d/Aiia_LC_B0034.jpg"> | ||
| + | <img alt="" src="https://static.igem.org/mediawiki/2011/8/8d/Aiia_LC_B0034.jpg" class="thumbimage" width="100%"></a></div></div> | ||
| + | </td></tr> | ||
| + | </table> | ||
| + | |||
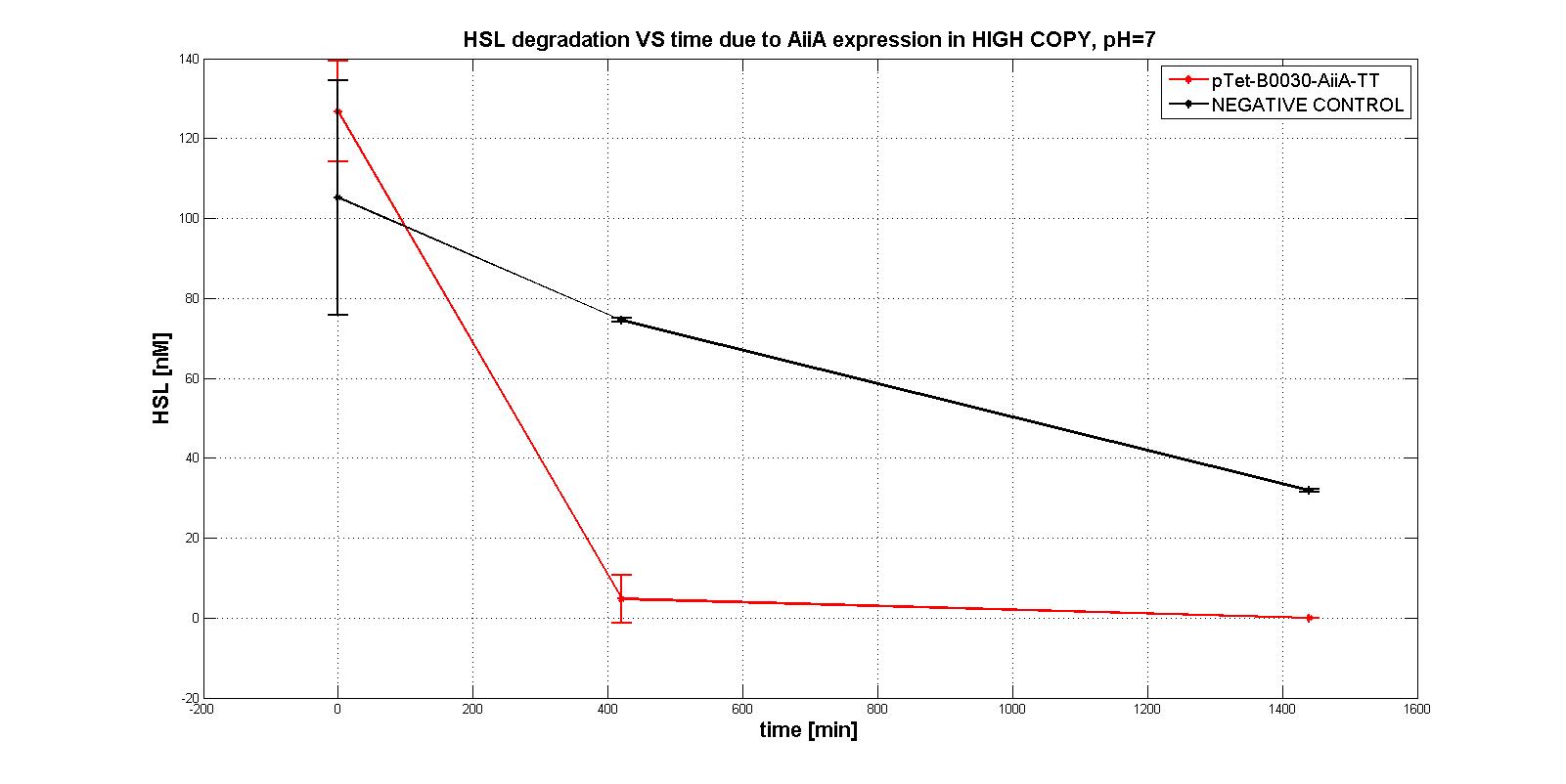
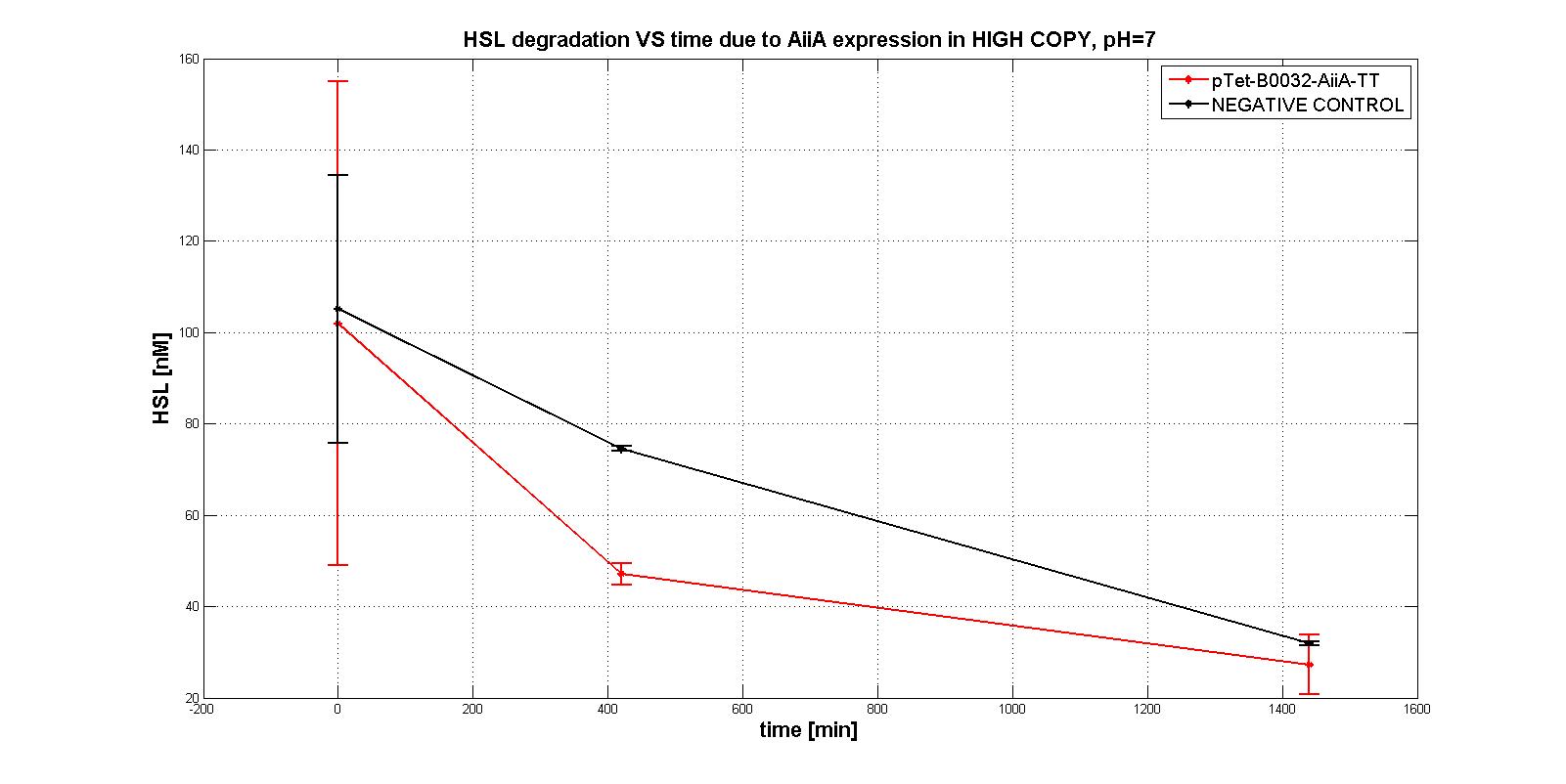
| + | We were only able to give semi-quantitative information about AiiA enzyme activity, performing an experiment in <em>E. coli</em> TOP10, with the four measurement devices in <a href="https://parts.igem.org/Part:pSB1A2">pSB1A2</a>; the results are summerized in the graphs below: | ||
| + | |||
| + | <table width='100%'><tr><td width='50%'> | ||
| + | <div style="text-align:justify"><div class="thumbinner" width='100%'> | ||
| + | <a href="https://static.igem.org/mediawiki/2011/6/6c/Aiia_HC_B0030.jpg"> | ||
| + | <img alt="" src="https://static.igem.org/mediawiki/2011/6/6c/Aiia_HC_B0030.jpg" class="thumbimage" width="100%"></a></div></div> | ||
| + | </td> | ||
| + | <td width='50%'> | ||
| + | <div style="text-align:justify"><div class="thumbinner" width='100%'> | ||
| + | <a href="https://static.igem.org/mediawiki/2011/e/ec/Aiia_HC_B0031.jpg"> | ||
| + | <img alt="" src="https://static.igem.org/mediawiki/2011/e/ec/Aiia_HC_B0031.jpg" class="thumbimage" width="100%"></a></div></div> | ||
| + | </td></tr> | ||
| + | <tr><td width='50%'> | ||
| + | <div style="text-align:justify"><div class="thumbinner" width='100%'> | ||
| + | <a href="https://static.igem.org/mediawiki/2011/b/b2/Aiia_HC_B0032.jpg"> | ||
| + | <img alt="" src="https://static.igem.org/mediawiki/2011/b/b2/Aiia_HC_B0032.jpg" class="thumbimage" width="100%"></a></div></div> | ||
| + | </td> | ||
| + | <td width='50%'> | ||
| + | <div style="text-align:justify"><div class="thumbinner" width='100%'> | ||
| + | <a href="https://static.igem.org/mediawiki/2011/d/d7/Aiia_HC_B0034.jpg"> | ||
| + | <img alt="" src="https://static.igem.org/mediawiki/2011/d/d7/Aiia_HC_B0034.jpg" class="thumbimage" width="100%"></a></div></div> | ||
| + | </td></tr> | ||
| + | </table> | ||
| + | |||
| + | In this way we can give a measure of AiiA activity as the percentage of HSL degraded in 7 hours (measurements provided as average [stdandard deviation]): | ||
| + | |||
| + | <div align="center"> | ||
| + | <table class="data"> | ||
| + | |||
| + | <tr><td class="row"><b>Measurement device</b></td><td><b> HSL degraded percentage </b></td></tr> | ||
| + | <tr><td class="row">BBa_K516220 </td><td class="row"> 96.11 [3.66]</td></tr> | ||
| + | <tr><td class="row">BBa_K516221 </td><td class="row"> 77.60 [2.66]</td></tr> | ||
| + | <tr><td class="row">BBa_K516222 </td><td class="row"> 50.66 [16.87]</td></tr> | ||
| + | <tr><td class="row">BBa_K516224</td><td class="row"> 100 [0]</td></tr> | ||
| + | </table></div> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | In order to evaluate the RBSs efficiency, we compute the ratio between the percentage of the degraded HSL realtive to the measurement device with the standard RBS (BBa_B0034): | ||
| + | <tr><td class="row"><b>RBS</b></td><td><b> RBS efficiency</b></td></tr> | ||
| + | <tr><td class="row">BBa_B0030 </td><td class="row"> 0.96</td></tr> | ||
| + | <tr><td class="row">BBa_B0031</td><td class="row"> 0.78</td></tr> | ||
| + | <tr><td class="row">BBa_B0032</td><td class="row"> 0.51</td></tr> | ||
| + | <tr><td class="row">BBa_B0034</td><td class="row"> 1</td></tr> | ||
| + | </table></div> | ||
Revision as of 14:16, 23 September 2011
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_C0060
User Reviews
UNIQ34e061e5dd431229-partinfo-00000000-QINU
|
•••••
UNIPV-Pavia iGEM 2011 |
NB: unless differently specified, all tests were performed in E. coli MGZ1 in M9 supplemented medium at 37°C in low copy plasmid pSB4C5.
A system of differential equations has been derived and we tried to identify the parameters, studying the exponential growth phase: The four measurment parts ptet-RBSx-LuxI were quantitatively characetrized using the BBa_T9002 biosensor. Unfortunately we were not able to estimate the model parameters (kcat, kM,AiiA) because placing the device in low copy plasmid (pSB4C5) we did not observe HSL degradation, as you can see in the figures below:
In order to evaluate the RBSs efficiency, we compute the ratio between the percentage of the degraded HSL realtive to the measurement device with the standard RBS (BBa_B0034): | ||||||||||||||||||
| RBS | RBS efficiency | ||||||||||||||||||
| BBa_B0030 | 0.96 | ||||||||||||||||||
| BBa_B0031 | 0.78 | ||||||||||||||||||
| BBa_B0032 | 0.51 | ||||||||||||||||||
| BBa_B0034 | 1 |
UNIQ34e061e5dd431229-partinfo-00000003-QINU