Difference between revisions of "Part:BBa K525121"
JSchwarzhans (Talk | contribs) (→Important parameters) |
(→Important parameters) |
||
| Line 9: | Line 9: | ||
===Important parameters=== | ===Important parameters=== | ||
| + | <center> | ||
| + | {|{{Table}} | ||
| + | !Experiment | ||
| + | !Characteristic | ||
| + | !Result | ||
| + | |- | ||
| + | |rowspan="6"|Expression | ||
| + | |- | ||
| + | |Compatibility | ||
| + | |''E. coli'' KRX | ||
| + | |- | ||
| + | |Inductor | ||
| + | |L-rhamnose | ||
| + | |- | ||
| + | |Optimal temperature | ||
| + | |37 °C | ||
| + | |- | ||
| + | |Specific growth rate (un-/induced) | ||
| + | |0.260 h<sup>-1</sup> / 0.106 h<sup>-1</sup> | ||
| + | |- | ||
| + | |Doubling time (un-/induced) | ||
| + | |2,67 h / 6.52 h | ||
| + | |- | ||
| + | |rowspan="5"|characteristics | ||
| + | |- | ||
| + | |Number of amino acids | ||
| + | | | ||
| + | |- | ||
| + | |Molecular weight | ||
| + | | | ||
| + | |- | ||
| + | |Theoretical pI | ||
| + | | | ||
| + | |- | ||
| + | |Localisation | ||
| + | |cell membrane | ||
| + | |- | ||
| + | |} | ||
| + | </center> | ||
| + | |||
<center> | <center> | ||
{|{{Table}} | {|{{Table}} | ||
Revision as of 21:07, 21 September 2011
S-layer cspB from Corynebacterium glutamicum with TAT-Sequence and lipid anchor, PT7 and RBS
Usage and Biology
S-layer proteins can be used as scaffold for nanobiotechnological applications and devices by e.g. fusing the S-layer's self-assembly domain to other functional protein domains. It is possible to coat surfaces and liposomes with S-layers. A big advantage of S-layers: after expressing in E. coli and purification, the nanobiotechnological system is cell-free. This enhances the biological security of a device.
This fluorescent S-layer fusion protein is used to characterize purification methods and the S-layer's ability to self-assemble on surfaces.
Important parameters
| Experiment | Characteristic | Result |
|---|---|---|
| Expression | ||
| Compatibility | E. coli KRX | |
| Inductor | L-rhamnose | |
| Optimal temperature | 37 °C | |
| Specific growth rate (un-/induced) | 0.260 h-1 / 0.106 h-1 | |
| Doubling time (un-/induced) | 2,67 h / 6.52 h | |
| characteristics | ||
| Number of amino acids | ||
| Molecular weight | ||
| Theoretical pI | ||
| Localisation | cell membrane |
| Experiment | Characteristic | Result |
|---|---|---|
| Expression (E. coli) | Localisation | cell membrane |
| Compatibility | E. coli KRX | |
| Inductor for expression | L-rhamnose for induction of T7 polymerase | |
| Specific growth rate (un-/induced) | 0.260 h-1 / 0.106 h-1 | |
| Doubling time (un-/induced) | 2.67 h / 6.52 h | |
| Characteristics | Molecular weight | |
| Theoretical pI |
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1421
Illegal XhoI site found at 248
Illegal XhoI site found at 866 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1400
Illegal SapI site found at 647
Illegal SapI site found at 859
Illegal SapI site found at 1407
Expression in E. coli
For characterization the CspB gen was fused with a monomeric RFP (BBa_E1010) using Gibson assembly.
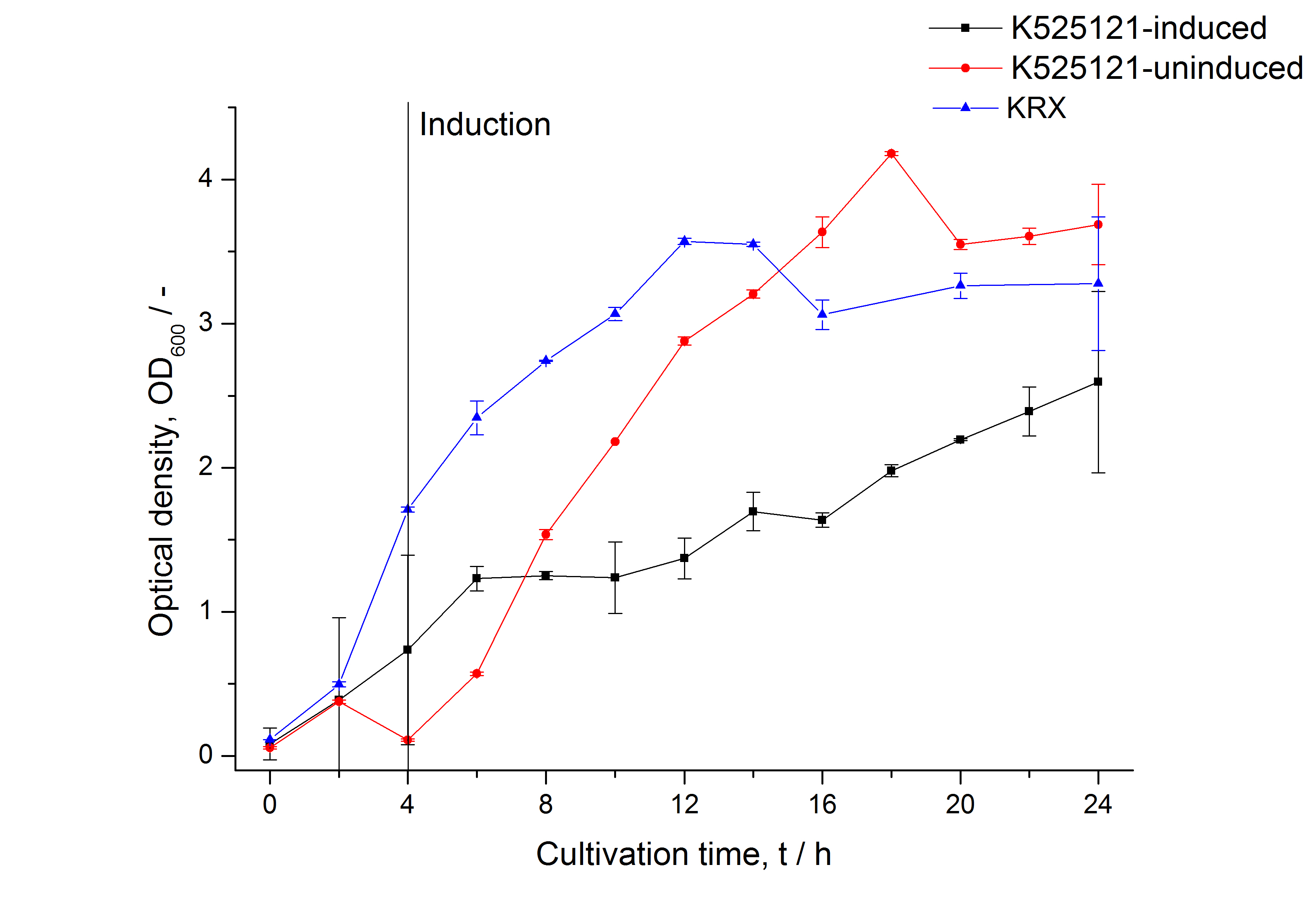
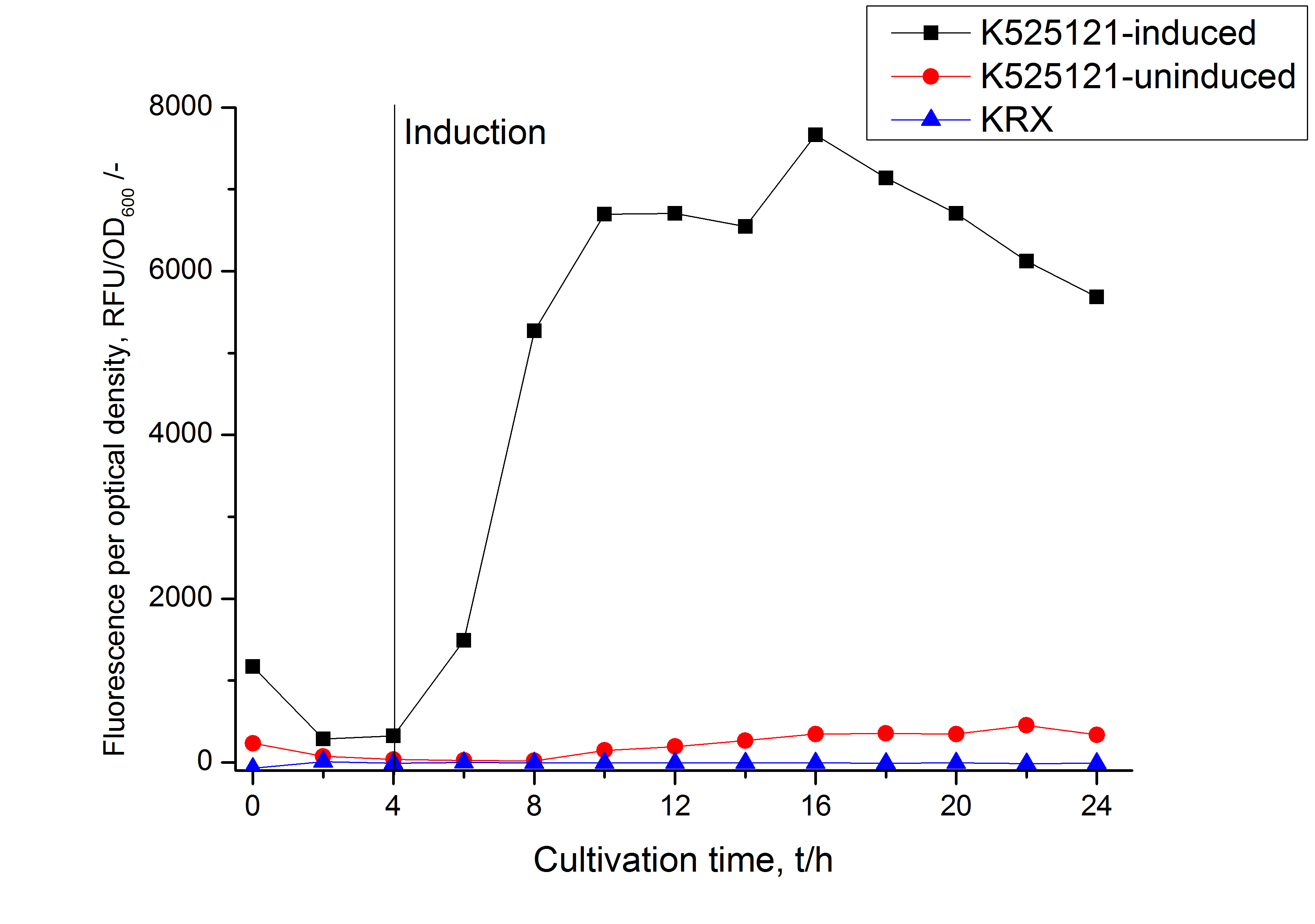
The CspB|mRFP fusion protein was overexpressed in E. coli KRX after induction of T7 polymerase by supplementation of 0,1 % L-rhamnose using the autinduction protocol from promega.
Identification and localisation
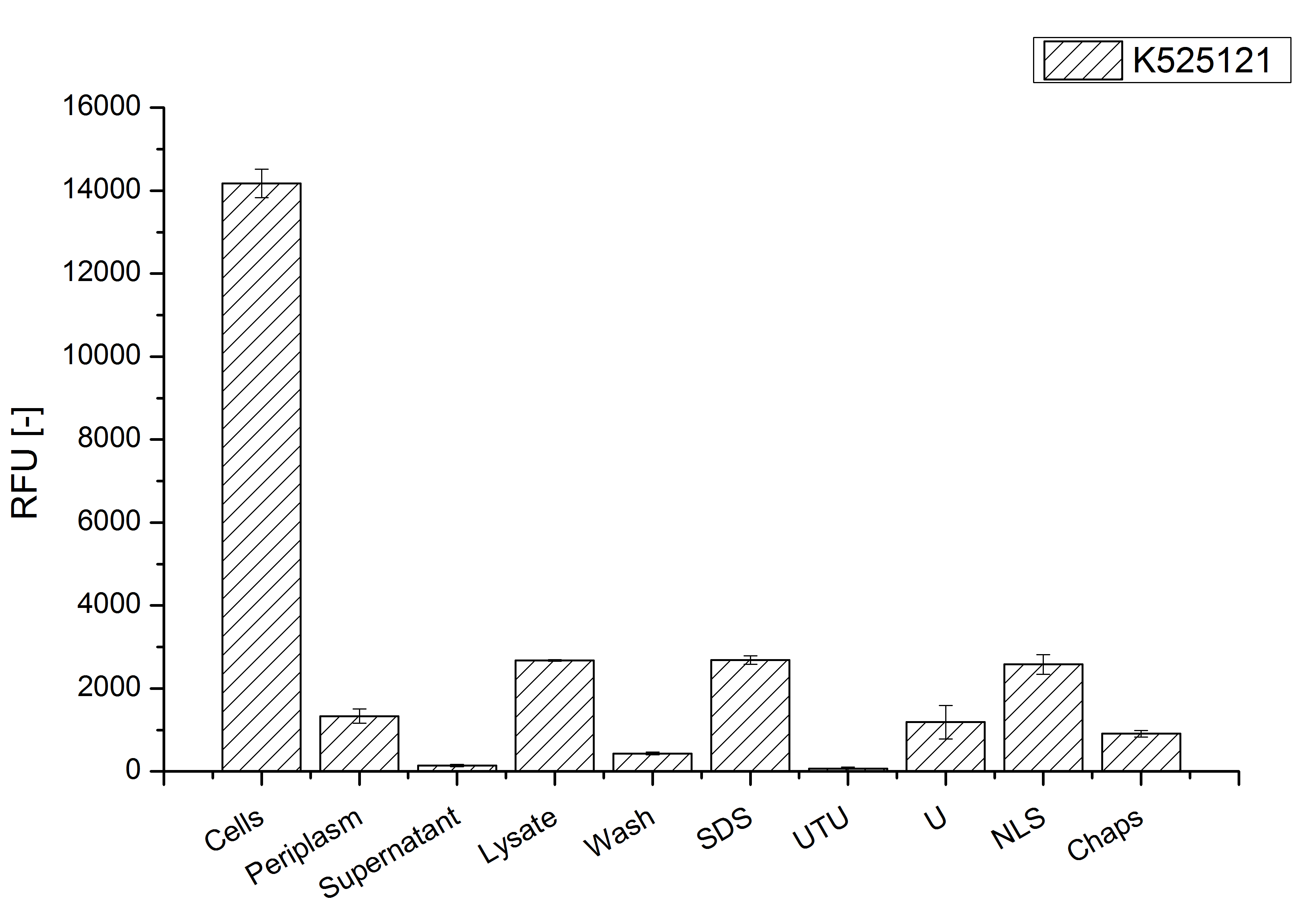
After a cultivation time of 18 h the CspB|mRFP fusion protein has to be localized in E. coli KRX. Therefore a part of the produced biomass was mechanically disrupted and the resulting lysate was washed with ddH2O. The periplasm was detached by using a osmotic shock from another part of the cells. The existance of fluorescene in the periplasm fraction, showed in fig. 3, indicates that Brevibacterium flavum TAT-signal sequence is at least in part functional in E. coli KRX.
The S-layer fusion protein could not be found in the polyacrylamide gel after a SDS-PAGE of the lysate and the cell depris were still red. This indicated that the fusion protein intigrates into the cell membrane with its lipid anchor. For testing this assumption the washed lysate was treted with ionic, nonionic and zwitterionic detergents to release the CspB|mRFP out of the membranes.
The existance of flourescence in the detergent fractions and the proportionally low fluorescence in the wash fraction confirm the hypothesis an insertion into the cell membrane (fig. 3). An insertion of these S-layer proteins might stabilize the membrane structure and increase the stability of cells against mechanical and chemical treatment. A stabilization of E. coli expressing S-lyer proteins was discribed by Lederer et al., (2010).
An other important fact is, that there is actually mRFP fluorescence measurable in such high concentrated detergent solutions. The S-layer seems to stabilize the biologically active conformation of mRFP. The MALDI-TOF analysis of the relevant size range in the polyacrylamid gel approved the existance of the intact fusion protein in all detergent fractions (fig x).
MALDI TOF analysis was first used to identify the location of the fusion protein in different fractions. Fractions of media supernatant after cultivation, periplasmatic isolation, cell lysis, denaturation in 6 M urea and the following wash with 2 % (v/v) Triton X-100, 2 % SDS (w/v) were loaded onto an SDS-PAGE and fragments of the gele were measured with MALDI TOF.