Difference between revisions of "Part:BBa K525222"
JSchwarzhans (Talk | contribs) |
JSchwarzhans (Talk | contribs) (→Important parameters) |
||
| Line 12: | Line 12: | ||
|rowspan="5"|[[Part:BBa_K525222#Expression_in_E._coli | Expression (''E. coli'')]] | |rowspan="5"|[[Part:BBa_K525222#Expression_in_E._coli | Expression (''E. coli'')]] | ||
|Localisation | |Localisation | ||
| − | | | + | |Cell membrane |
|- | |- | ||
|Compatibility | |Compatibility | ||
| − | |''E. coli'' KRX | + | |''E. coli'' KRX |
|- | |- | ||
|Induction of expression | |Induction of expression | ||
| − | | | + | |L-rhamnose for induction of T7 polymerase |
|- | |- | ||
|Specific growth rate (un-/induced) | |Specific growth rate (un-/induced) | ||
| Line 41: | Line 41: | ||
<partinfo>BBa_K525222 parameters</partinfo> | <partinfo>BBa_K525222 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
===Expression in ''E. coli''=== | ===Expression in ''E. coli''=== | ||
The CspB gen was fused with a monomeric RFP ([https://parts.igem.org/Part:BBa_E1010 BBa_E1010]) using [http://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly] for characterization. | The CspB gen was fused with a monomeric RFP ([https://parts.igem.org/Part:BBa_E1010 BBa_E1010]) using [http://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly] for characterization. | ||
Revision as of 21:07, 21 September 2011
S-layer cspB from Corynebacterium halotolerans
S-layer Corynebacterium halotolerans
Important parameters
| Experiment | Characteristic | Result |
|---|---|---|
| Expression (E. coli) | Localisation | Cell membrane |
| Compatibility | E. coli KRX | |
| Induction of expression | L-rhamnose for induction of T7 polymerase | |
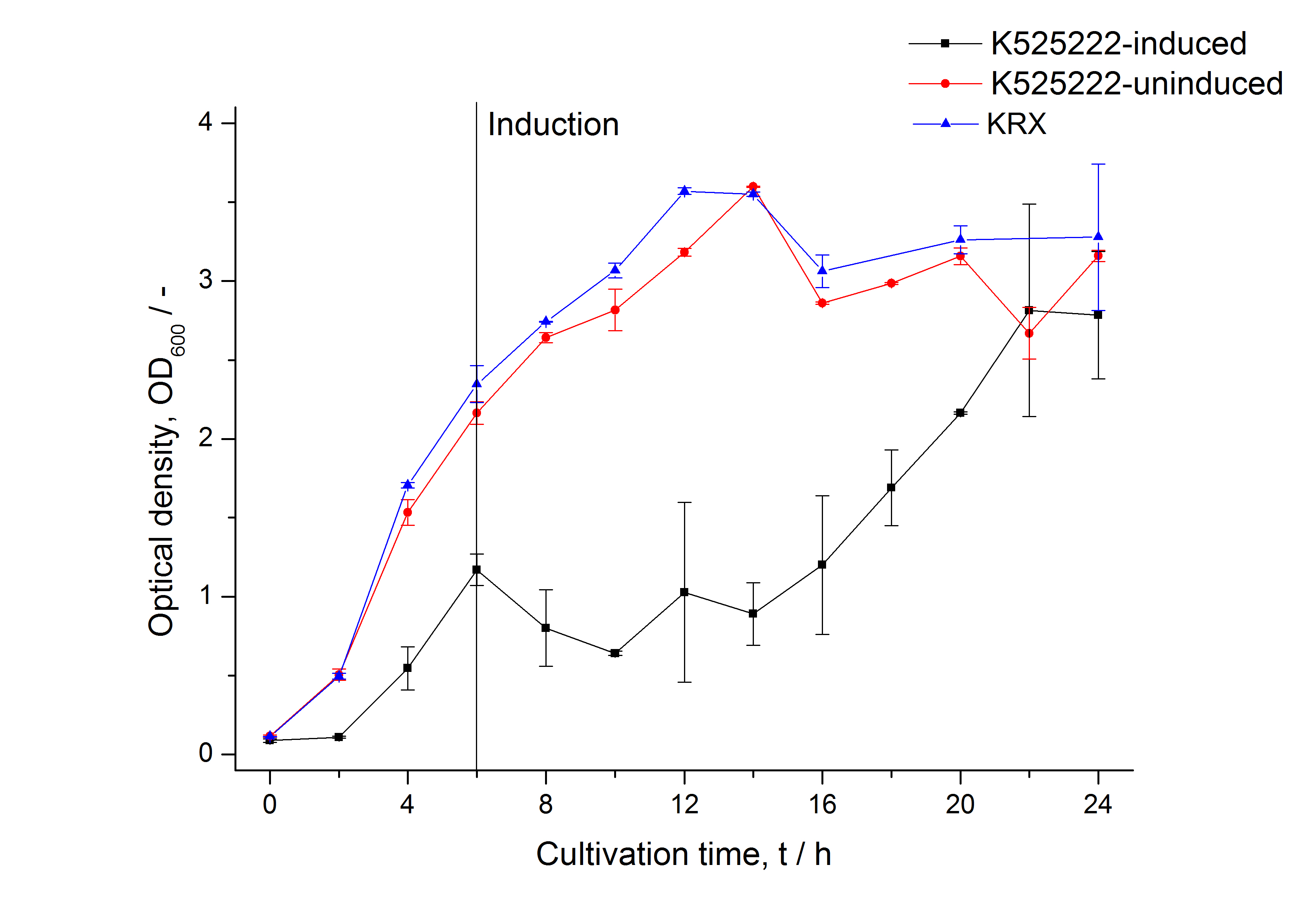
| Specific growth rate (un-/induced) | 0.245 h-1 / 0.109 h-1 | |
| Doubling time (un-/induced) | 2.83 h / 6.33 h |
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1103
Illegal XhoI site found at 559 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 226
Illegal NgoMIV site found at 1315
Illegal AgeI site found at 217
Illegal AgeI site found at 458
Illegal AgeI site found at 505 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 907
Illegal BsaI.rc site found at 214
Illegal BsaI.rc site found at 592
Illegal BsaI.rc site found at 994
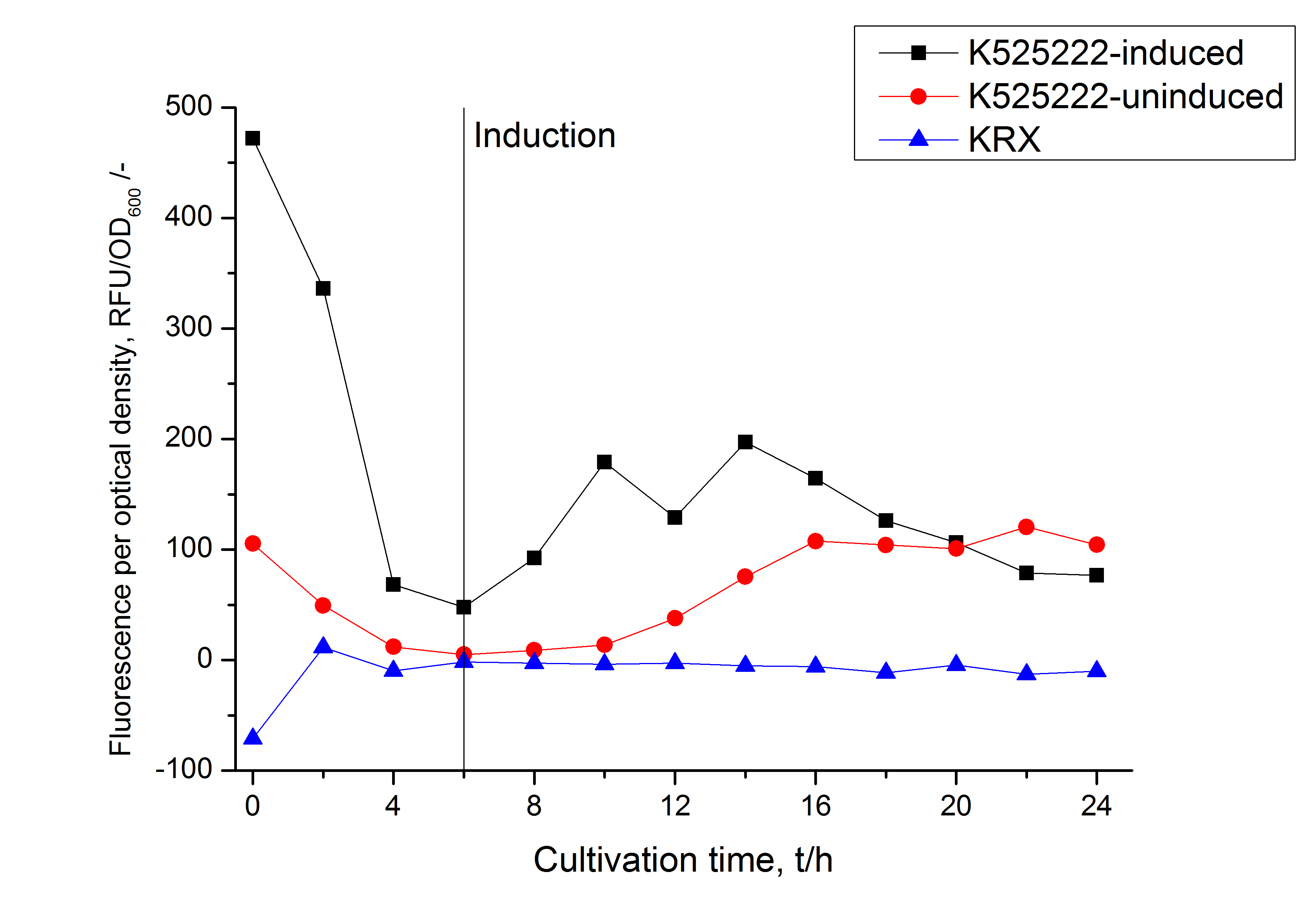
Expression in E. coli
The CspB gen was fused with a monomeric RFP (BBa_E1010) using [http://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly] for characterization.
The CspB|mRFP fusion protein was overexpressed in E. coli KRX after induction of T7 polymerase by supplementation of 0,1 % L-rhamnose using the [http://2011.igem.org/Team:Bielefeld-Germany/Protocols/Downstream-processing#Expression_of_S-layer_genes_in_E._coli autinduction protocol] from promega.