Difference between revisions of "Part:BBa K638301"
| Line 14: | Line 14: | ||
<gallery widths=400px heights=500px> | <gallery widths=400px heights=500px> | ||
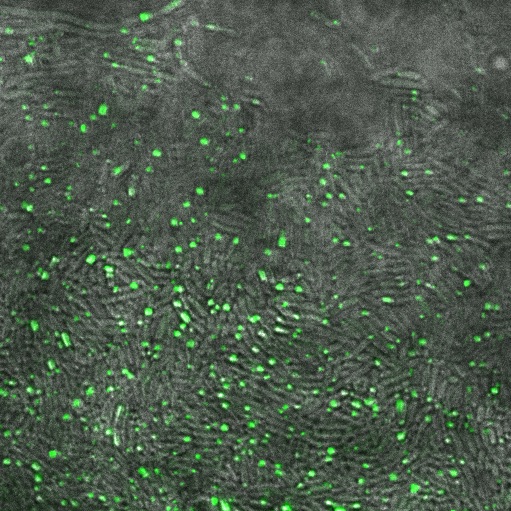
Image:Cam_Reflectin-GFP-inclusionbodies.jpg | Reflectin-GFP on [[Part:pSB1A3 | pSB1A3]](a high copy plasmid), induced with 1mM Arabinose. Fluorescence is localised in dots at the end of the cells, indicating that fusion protein is localised to insoluble inclusion bodies. | Image:Cam_Reflectin-GFP-inclusionbodies.jpg | Reflectin-GFP on [[Part:pSB1A3 | pSB1A3]](a high copy plasmid), induced with 1mM Arabinose. Fluorescence is localised in dots at the end of the cells, indicating that fusion protein is localised to insoluble inclusion bodies. | ||
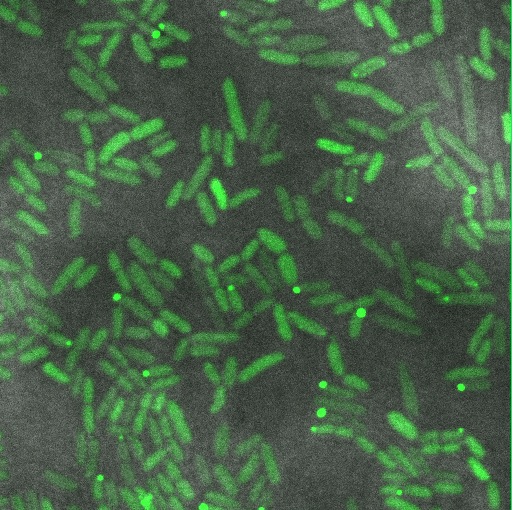
| − | Image:Cam-Low-copy-reflectin.jpg | Reflectin-GFP on [[Part:pSB3k3 | pSB3K3]] (low/med copy plasmid), induced with 1mM Arabinose. The fusion protein is distributed throughout the cytoplasm, indicating soluble protein is expressed. | + | Image:Cam-Low-copy-reflectin.jpg | Reflectin-GFP on [[Part:pSB3k3 | pSB3K3]] (low/med copy plasmid), induced with 1mM Arabinose. The fusion protein is distributed throughout the cytoplasm, indicating soluble protein is expressed. |
</gallery> | </gallery> | ||
| − | From this confocal imaging, we recommend that reflectin be expressed on a low/med copy number backbone to allow protein folding to occur to give soluble protein. High copy number plasmids give inclusion bodies. | + | From this confocal imaging, we recommend that reflectin be expressed on a '''low/med copy number backbone''' to allow protein folding to occur to give soluble protein. High copy number plasmids give inclusion bodies. Some inclusion bodies are also visible in the low copy construct, suggesting that some cells are expressing more fusion protein than others. See our [[Part:BBa_I0500:Experience | experience]] of the pBAD promoter for a discussion of variations of induction within a population of cells. |
Revision as of 19:23, 21 September 2011
Arabinose inducible Reflectin A1-sfGFP fusion generator
E.coli transformed with this construct will produce a superfolder Green fluorescent Protein (sfGFP) fused variant of reflectin A1 under the control of pBAD, an arabinose inducible protein. There is currently (at the time of writing) no good assay for reflectin, in vivo. This construct allows reflectin to be localised within the cell. This part has been used to view reflectin containing inclusion bodies in E.coli and the sfGFP retained its fluorescence despite the fusion.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1205
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1144
Illegal BamHI site found at 1240 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 979
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 961
Illegal SapI.rc site found at 2307
Usage and Biology
Cambridge 2011 used this part to assess reflectin folding on different backbones.
Reflectin-GFP on pSB1A3(a high copy plasmid), induced with 1mM Arabinose. Fluorescence is localised in dots at the end of the cells, indicating that fusion protein is localised to insoluble inclusion bodies.
Reflectin-GFP on pSB3K3 (low/med copy plasmid), induced with 1mM Arabinose. The fusion protein is distributed throughout the cytoplasm, indicating soluble protein is expressed.
From this confocal imaging, we recommend that reflectin be expressed on a low/med copy number backbone to allow protein folding to occur to give soluble protein. High copy number plasmids give inclusion bodies. Some inclusion bodies are also visible in the low copy construct, suggesting that some cells are expressing more fusion protein than others. See our experience of the pBAD promoter for a discussion of variations of induction within a population of cells.