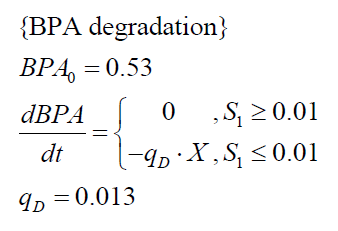Difference between revisions of "Part:BBa K525517"
(→Important parameters) |
(→Important parameters) |
||
| Line 43: | Line 43: | ||
|450 nm (due to CYP) | |450 nm (due to CYP) | ||
|- | |- | ||
| − | |rowspan="2"| | + | |rowspan="2"|Degradation of BPA |
|[[Part:BBa_K525517#Bisphenol_A_degradation_with_E._coli | Completely degradation of 0.53 mM BPA]] | |[[Part:BBa_K525517#Bisphenol_A_degradation_with_E._coli | Completely degradation of 0.53 mM BPA]] | ||
|21 - 24 h | |21 - 24 h | ||
Revision as of 14:16, 17 September 2011
Fusion protein of BisdA and BisdB behind constitutive promoter
Fusion protein of BisdA and BisdB behind constitutive promoter leading to enhanced bisphenol A degradation compared to polycistronic bisdAB gene.
Usage and Biology
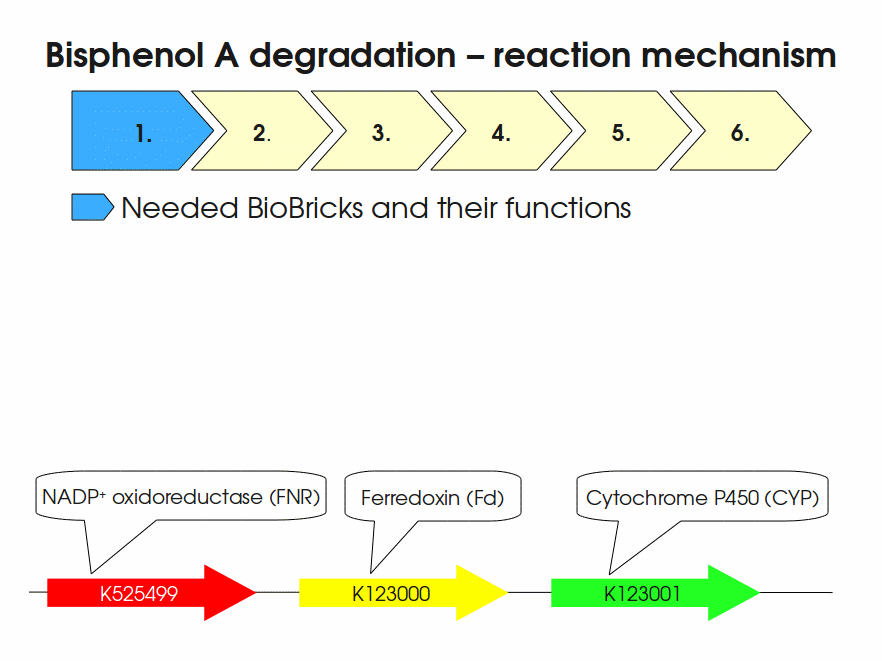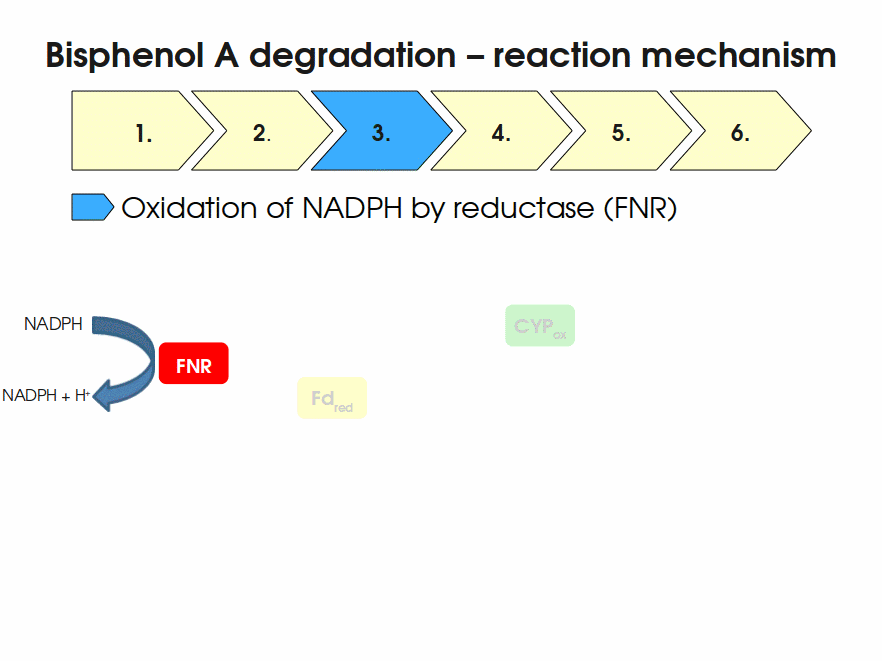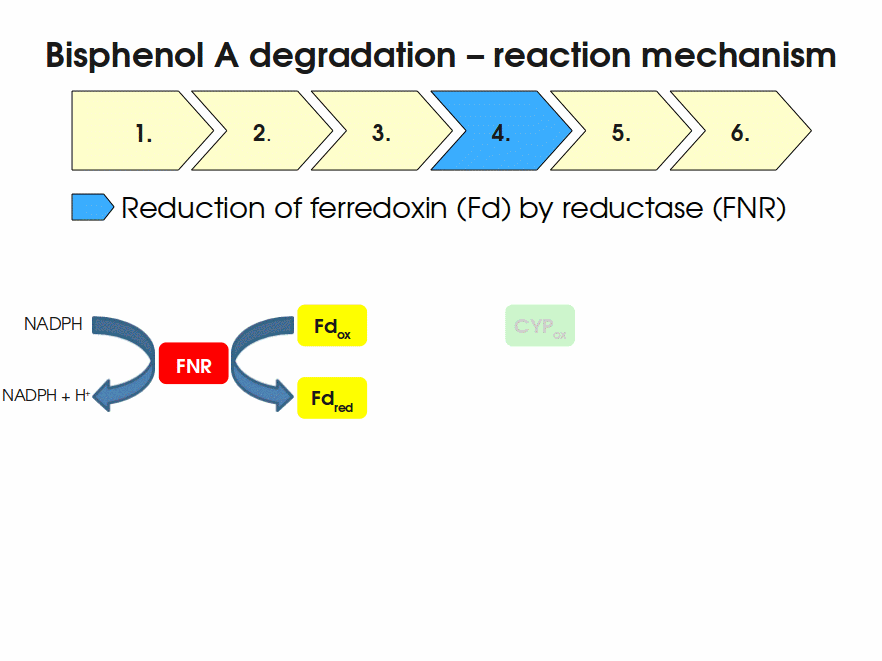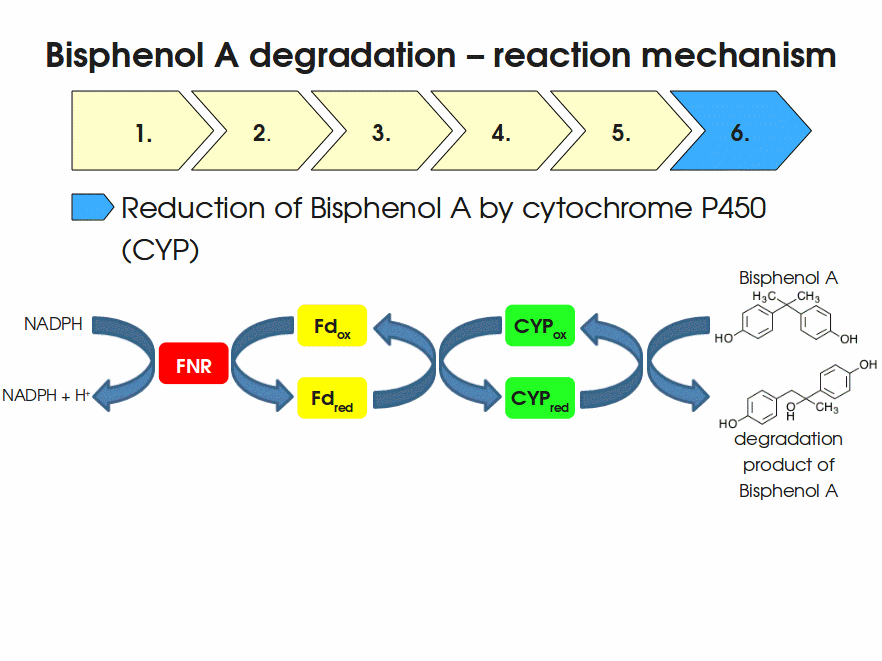
Expressing this BioBrick in E. coli enables the bacterium to degrade the endocrine disruptor bisphenol A (BPA). BPA is mainly hydroxylated into the products 1,2-Bis(4-hydroxyphenyl)-2-propanol and 2,2-Bis(4-hydroxyphenyl)-1-propanol. In S. bisphenolicum AO1, a total of three genes are responsible for this BPA hydroxylation: a cytochrome P450 (CYP, bisdB), a ferredoxin (Fd, bisdA) and a ferredoxin-NAD+ oxidoreductase (FNR) Sasaki05a. The three gene products act together to reduce BPA while oxidizing NADH + H+. The cytochrome P450 (BisdB) reduces the BPA and is oxidized during this reaction. BisdB in its oxidized status is reduced by the ferredoxin (BisdA) so it can reduce BPA again. The oxidized BisdA is reduced by a ferredoxin-NAD+ oxidoreductase consuming NADH + H+ so the BPA degradation can continue Sasaki05a. This electron transport chain between the three enzymes involved in BPA degradation and the BioBricks needed to enable this reaction in vivo and in vitro are shown in the following figure (please have some patience, it's an animated .gif file):
Important parameters
| Experiment | Characteristic | Result |
|---|---|---|
| Expression in E. coli | Compatibility | E. coli KRX, TOP10, MACH1, BL21(DE3) |
| Expression | Constitutive | |
| Optimal temperature | 30 °C | |
| BPA working concentration | 120 mg L-1 (0.53 mM) | |
| Purification | Molecular weight | 59.3 kDa |
| Theoretical pI | 4.99 | |
| High absorbtion | 450 nm (due to CYP) | |
| Degradation of BPA | Completely degradation of 0.53 mM BPA | 21 - 24 h |
| Specific BPA degradation rate | 1.29 10-10 mM cell-1 |
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 664
Illegal BamHI site found at 1402 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 65
Illegal AgeI site found at 1655 - 1000COMPATIBLE WITH RFC[1000]
Bisphenol A degradation with E. coli
The bisphenol A degradation with the BioBricks BBa_K123000 and BBa_K123001 works in E. coli KRX in general. Because [http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2672.2008.03843.x/full Sasaki et al. (2008)] reported problems with protein folding in E. coli which seem to avoid a complete BPA degradation, we did not cultivate at 37 °C and we did not use the strong T7 promoter as [http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2672.2008.03843.x/full Sasaki et al. (2008)] did for expressing these BioBricks but we cultivated at 30 °C and we used a medium strong constitutive promoter (BBa_J23110). 30 °C is in addition the cultivation temperature of S. bisphenolicum AO1. With this promoter upstream of a polycistronic bisdAB gene we were able to completely degrade 120 mg L-1 BPA in about 30 - 33 h. By fusing BBa_K123000 and BBa_K123001 together (using Freiburg BioBrick assembly standard) we could improve the BPA degradation of E. coli even further, so 120 mg L-1 BPA can be degraded in 21 - 24 h. This data is shown in the following figure:
We also carried out these cultivations at different temperatures and BPA concentrations, but the chosen conditions (30 °C and 120 mg L-1 BPA) seem to be the best. Higher BPA concentrations have an effect on the growth of E. coli and higher temperature leeds to a worse BPA degradation (probably due to misfolding of the enzymes). Lower temperature also leeds to less BPA degradation (probably due to slower growth, expression and reaction rate at lower temperatures). These data on the effect of the temperature on the BPA degradation is shown in fig. 3.
As shown by [http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2672.2008.03843.x/full Sasaki et al. (2008)], BisdB expressed in E. coli leeds to hardly no BPA degradation. In our experiments we could not detect the BPA degradation products 1,2-Bis(4-hydroxyphenyl)-2-propanol and 2,2-Bis(4-hydroxyphenyl)-1-propanol in cultivations with E. coli expressing BBa_K123000 or BBa_K123001 alone (neither via UV- nor MS-detection). The BPA degradation products 1,2-Bis(4-hydroxyphenyl)-2-propanol and 2,2-Bis(4-hydroxyphenyl)-1-propanol were identified via MS-MS (m/z: 243 / 225 / 211 / 135) and only occured in cultivations with E. coli expressing BisdA and BisdB together. [http://www.springerlink.com/content/q7864l02734wg32m/ Sasaki et al. (2005)] reported the same MS-MS results for 1,2-Bis(4-hydroxyphenyl)-2-propanol and 2,2-Bis(4-hydroxyphenyl)-1-propanol when degrading BPA with S. bisphenolicum AO1 as we observed in our BPA degradation experiments.
We could also identify the BPA degradation products when working with E. coli TOP10 and MACH1 (data not shown) but because we want to fuse BisdA and BisdB to S-layer proteins which we express in E. coli KRX the characterizations of BBa_K123000 and BBa_K123001 were carried out in this strain.
Modelling of intracellular bisphenol A degradation
The modelling was done with the software [http://www.berkeleymadonna.com/ Berkeley Madonna] using the [http://en.wikipedia.org/wiki/Runge–Kutta_methods#Common_fourth-order_Runge.E2.80.93Kutta_method common fourth-order Runge-Kutta] method to solve the equations. The model was fitted to the measured data shown above by the function "curve fit" in Berkeley Madonna to calculate the parameters, constants etc.
To model the BPA degradation by E. coli carrying BioBricks for BPA degradation (BBa_K123000 and BBa_K123001) the cell growth has to be described first to calculate a specific BPA degradation rate per cell. The observed growth of E. coli on (our) LB medium was [http://en.wikipedia.org/wiki/Diauxie diauxic] with two different growth phases. Cell growth is a [http://en.wikipedia.org/wiki/First_order_kinetics#First-order_reactions first-order reaction] and is mathematically described as
with the specific growth rate µ and the cell count X. The specific growth rate is dependent on the concentration of the growth limiting substrate (e.g. glucose) and can be described as
with the substrate concentration S, the Monod constant KS and the maximal specific growth rate µmax ([http://www.annualreviews.org/doi/abs/10.1146/annurev.mi.03.100149.002103 Monod, 1949]). Because LB medium is a complex medium we cannot measure the substrate concentration so we have to assume an imaginary substrate concentration. Due to the diauxic growth two different substrates with different Monod constants and consumption rates are necessary to model the cell growth. The amount of a substrate S can be modelled as follows
with the specific substrate consumption rate per cell qS. The whole model for the diauxic growth of E. coli on LB medium with two not measurable (imaginary) substrates looks like:
The specific BPA degradation rate per cell qD is modelled with an equation like eq. (3). In the beginning of the cultivations, when E. coli growths on the "good" imaginary substrate S1, no BPA degradation is observed. When this substrate is consumed, the BPA degradation starts. The model for this diauxic behavior is as follows:
Fig. 4 shows a comparison between modelled and measured data for cultivations with BPA degrading E. coli. In Tab. 2 the parameters for the model are given, obtained by curve fitting the model to the data.

Tab. 2: Parameters of the model.
| Parameter | BBa_K525512 | BBa_K525517 |
|---|---|---|
| X0 | 0.112 108 mL-1 | 0.138 108 mL-1 |
| µmax | 1.253 h-1 | 1.357 h-1 |
| KS,1 | 2.646 AU-1 | 1.92 AU-1 |
| KS,2 | 265.1 AU-1 | 103.1 AU-1 |
| S1,0 | 1.688 AU | 1.166 AU |
| qS,1 | 0.478 AU 10-8 cell-1 | 0.319 AU 10-8 cell-1 |
| S2,0 | 16.091 AU | 6.574 AU |
| qS,2 | 0.295 AU 10-8 cell-1 | 0.191 AU 10-8 cell-1 |
| BPA0 | 0.53 mM | 0.53 mM |
| qD | 8.76 10-11 mM cell-1 | 1.29 10-10 mM cell-1 |
The specific BPA degradation rate per cell qD is about 50 % higher when using the fusion protein compared to the polycistronic bisdAB gene. This results in an average 9 hours faster, complete BPA degradation by E. coli carrying BBa_K525517 compared to BBa_K525512 as observed during our cultivations. The fusion protein between BisdA and BisdB improves the BPA degradation by E. coli.
Interpretation of the results
Degrading BPA with E. coli using the BisdA | BisdB fusion protein, both domains (BisdA and BisdB) are active and correctly folded because otherwise there would be no BPA degradation measured and no BPA degradation products 1,2-Bis(4-hydroxyphenyl)-2-propanol and 2,2-Bis(4-hydroxyphenyl)-1-propanol detected which could definetely be identified via MS-MS. The about 50 % higher specific BPA degradation rate in E. coli expressing BisdA | BisdB fusion protein could be explained either by improved folding properties of the fusion protein or by the closer distance of BisdA and BisdB in the fusion protein leeding to a more efficient reaction.
Methods
Cultivations

- Chassis: Promega's [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ E. coli KRX]
- Medium: LB medium supplemented with 100 mg L-1 Ampicillin and 120 mg L-1 bisphenol A (Sigma, 97 %)
- BPA is thermally stable -> you can autoclave it together with the medium
- 100 mL culture in 300 mL shaking flask without baffles (Schott) with silicon plugs
- Cultivation temperature: 24 °C, 30 °C or 37 °C, tempered with Infors AG AQUATRON at 120 rpm
- for characterizations: automatic sampling every three hours with Gilson fraction controller F2XX cooled (< 4 °C) with Julabo F10 water bath
- the characterization experiment setup is shown on the picture on the right
HPLC method
- C18 reverse phase column
- Isocratic method: 45 % Acetonitrile
- Flow = 0.6 mL min-1
- UV-detection at 227 nm
- Internal standard: 100 mg L-1 bisphenol F (BPF)
- Column:
- Eurospher II 100-5 C18p by [http://www.knauer.net/ Knauer]
- Dimensions: 150 x 4.6 mm with precolumn
- Particle size: 5 µm
- Pore size: 100 Å
- Material: silica gel
- Software:
- Clarity (Version 3.0.5.505) by [http://www.dataapex.com/ Data Apex]
- Autosampler:
- Midas by [http://www.spark.nl/ Spark Holland]
- Tray cooling: 10 °C
- Pump:
- L-6200A Intelligent Pump by [http://www.hitachi.com/ Hitachi]
- UV-Detector:
- Series 1050 by [http://www.hp.com/ Hewlett Packard]
References
Monod J (1949) The growth of bacterial cultures, Annu Rev Microbiol [http://www.annualreviews.org/doi/abs/10.1146/annurev.mi.03.100149.002103 3:371-394].
<biblio>
- Sasaki pmid=18492046
- Sasaki05a pmid=16332782
</biblio>