Difference between revisions of "Part:BBa K404122"
| Line 4: | Line 4: | ||
[[Image:Freiburg10_Vectorplasmid composite 4.png|thumb|center|480px]]<br> | [[Image:Freiburg10_Vectorplasmid composite 4.png|thumb|center|480px]]<br> | ||
<h2>ITRs</h2><br> | <h2>ITRs</h2><br> | ||
| − | + | <html> | |
| + | <center> | ||
| + | <img src=https://static.igem.org/mediawiki/2010/8/88/Freiburg10_organization_ITRs.png width=400> | ||
| + | </center> | ||
| + | </html><br> | ||
The inverted terminal repeat structures can be subdivided into several palindromic motives: A and A’ form a stem loop which encases B and B’ as well as C and C’. Those motives form both arms of the T-shaped structure. The functional motives on the ITR are two regions that bind Rep 68/78, called Rep-binding elements (RBE on the stem and RBE’ on the B arm) and the terminal resolution site (trs) in which the rep proteins introduce single-stranded nicks. The 3’ OH end of the A motive acts as a primer for DNA replication (Im & Muzyczka, 1990) (Lusby, Fife, & Berns, 1980). | The inverted terminal repeat structures can be subdivided into several palindromic motives: A and A’ form a stem loop which encases B and B’ as well as C and C’. Those motives form both arms of the T-shaped structure. The functional motives on the ITR are two regions that bind Rep 68/78, called Rep-binding elements (RBE on the stem and RBE’ on the B arm) and the terminal resolution site (trs) in which the rep proteins introduce single-stranded nicks. The 3’ OH end of the A motive acts as a primer for DNA replication (Im & Muzyczka, 1990) (Lusby, Fife, & Berns, 1980). | ||
<h2>CMV promoter</h2><br> | <h2>CMV promoter</h2><br> | ||
<h2>Beta-globin-intron</h2><br> | <h2>Beta-globin-intron</h2><br> | ||
| + | <html> | ||
| + | <center> | ||
| + | <img src=https://static.igem.org/mediawiki/2010/7/7c/Freiburg10_FACS_betaglobin.png width=400> | ||
| + | </center> | ||
| + | </html><br> | ||
| + | <html> | ||
| + | <center> | ||
| + | <img src=https://static.igem.org/mediawiki/2010/2/26/Freiburg10_Diagram_betaglobin.png.png width=400> | ||
| + | </center> | ||
| + | </html><br> | ||
| + | |||
| + | The beta-globin intron BioBrick consists of a partial chimeric CMV promoter followed by the intron II of the beta-globin gene. The 3´end of the intron is fused to the first 25 bases of human beta globin gene exon 3. The beta globin intron BioBrick is assumed to enhance eukaryotic gene expression (Nott, Meislin, & Moore, 2003). As shown in Figure 9 and Figure 10 the vectorplasmid missing the beta-globin intron showed a negligible difference in mVenus expression compared to viral genomes containing the beta-globin intron. Considering these results and taking into account that a constant volume of viral particles has been used for transduction, the difference between the construct containing and lacking the beta-globin intron is minimal. Since packaging efficiency of the AAV-2 decreases with increasing sizes of the insert (Dong, Fan, & Frizzell, 1996), the iGEM team Freiburg_Bioware suggests using the beta-globin intron in dependence on the size of your transgene. | ||
<h2>mGMK_TK30 fusion protein</h2><br> | <h2>mGMK_TK30 fusion protein</h2><br> | ||
<h2>hGH</h2><br> | <h2>hGH</h2><br> | ||
Revision as of 17:42, 26 October 2010
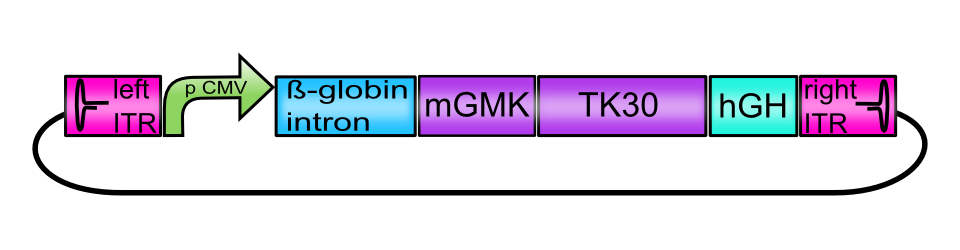
[AAV2]-left-ITR_pCMV_betaglobin_mGMK_TK30_hGH_[AAV2]-right-ITR
ITRs

The inverted terminal repeat structures can be subdivided into several palindromic motives: A and A’ form a stem loop which encases B and B’ as well as C and C’. Those motives form both arms of the T-shaped structure. The functional motives on the ITR are two regions that bind Rep 68/78, called Rep-binding elements (RBE on the stem and RBE’ on the B arm) and the terminal resolution site (trs) in which the rep proteins introduce single-stranded nicks. The 3’ OH end of the A motive acts as a primer for DNA replication (Im & Muzyczka, 1990) (Lusby, Fife, & Berns, 1980).
CMV promoter
Beta-globin-intron


The beta-globin intron BioBrick consists of a partial chimeric CMV promoter followed by the intron II of the beta-globin gene. The 3´end of the intron is fused to the first 25 bases of human beta globin gene exon 3. The beta globin intron BioBrick is assumed to enhance eukaryotic gene expression (Nott, Meislin, & Moore, 2003). As shown in Figure 9 and Figure 10 the vectorplasmid missing the beta-globin intron showed a negligible difference in mVenus expression compared to viral genomes containing the beta-globin intron. Considering these results and taking into account that a constant volume of viral particles has been used for transduction, the difference between the construct containing and lacking the beta-globin intron is minimal. Since packaging efficiency of the AAV-2 decreases with increasing sizes of the insert (Dong, Fan, & Frizzell, 1996), the iGEM team Freiburg_Bioware suggests using the beta-globin intron in dependence on the size of your transgene.
mGMK_TK30 fusion protein
hGH
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1319
Illegal NgoMIV site found at 1977
Illegal NgoMIV site found at 2882
Illegal AgeI site found at 3011 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 3413
Illegal SapI.rc site found at 1369