Difference between revisions of "Part:BBa K404122"
| Line 4: | Line 4: | ||
[[Image:Freiburg10_Vectorplasmid composite 4.png|thumb|center|480px]]<br> | [[Image:Freiburg10_Vectorplasmid composite 4.png|thumb|center|480px]]<br> | ||
<h2>ITRs</h2><br> | <h2>ITRs</h2><br> | ||
| + | [[Image:Freiburg10_ITR.png|thumb|center|480px]]<br> | ||
| + | The inverted terminal repeat structures can be subdivided into several palindromic motives: A and A’ form a stem loop which encases B and B’ as well as C and C’. Those motives form both arms of the T-shaped structure. The functional motives on the ITR are two regions that bind Rep 68/78, called Rep-binding elements (RBE on the stem and RBE’ on the B arm) and the terminal resolution site (trs) in which the rep proteins introduce single-stranded nicks. The 3’ OH end of the A motive acts as a primer for DNA replication (Im & Muzyczka, 1990) (Lusby, Fife, & Berns, 1980). | ||
<h2>CMV promoter</h2><br> | <h2>CMV promoter</h2><br> | ||
<h2>Beta-globin-intron</h2><br> | <h2>Beta-globin-intron</h2><br> | ||
Revision as of 16:47, 26 October 2010
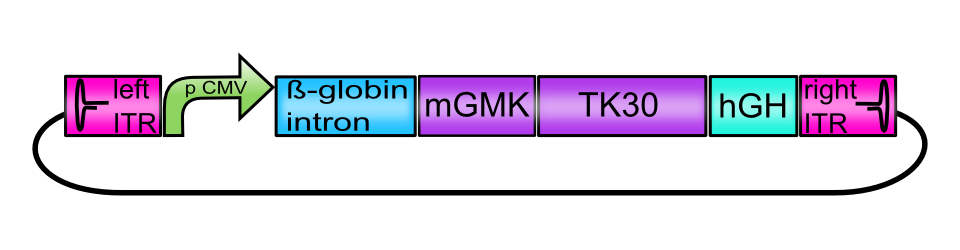
[AAV2]-left-ITR_pCMV_betaglobin_mGMK_TK30_hGH_[AAV2]-right-ITR
ITRs
The inverted terminal repeat structures can be subdivided into several palindromic motives: A and A’ form a stem loop which encases B and B’ as well as C and C’. Those motives form both arms of the T-shaped structure. The functional motives on the ITR are two regions that bind Rep 68/78, called Rep-binding elements (RBE on the stem and RBE’ on the B arm) and the terminal resolution site (trs) in which the rep proteins introduce single-stranded nicks. The 3’ OH end of the A motive acts as a primer for DNA replication (Im & Muzyczka, 1990) (Lusby, Fife, & Berns, 1980).
CMV promoter
Beta-globin-intron
mGMK_TK30 fusion protein
hGH
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1319
Illegal NgoMIV site found at 1977
Illegal NgoMIV site found at 2882
Illegal AgeI site found at 3011 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 3413
Illegal SapI.rc site found at 1369