Difference between revisions of "Part:BBa R0082"
| Line 11: | Line 11: | ||
In nature, this promoter is upstream of the ompC porin gene. The regulation of ompC is determined by the EnvZ-OmpR osmosensing machinery. EnvZ phosphorylates OmpR to OmpR-P. At high osmolarity, EnvZ is more active, creating more OmpR-P. OmpR-P then binds to the low-affinity OmpR operator sites upstream of ompC. | In nature, this promoter is upstream of the ompC porin gene. The regulation of ompC is determined by the EnvZ-OmpR osmosensing machinery. EnvZ phosphorylates OmpR to OmpR-P. At high osmolarity, EnvZ is more active, creating more OmpR-P. OmpR-P then binds to the low-affinity OmpR operator sites upstream of ompC. | ||
| + | ==Contribution: Hamburg 2016== | ||
| + | '''Group:''' Hamburg / '''Authors:''' Kai Pohl, Daniel Wedemeyer | ||
| + | |||
| + | '''Summary:''' We analysed the noise produced by BBa_R0082 by flow cytometry using a ΔEnvZ strain. | ||
| + | |||
| + | <p>In two approaches, we ligated BBa_R0082 (OmpR, positive promoter) with BBa_E0240 (GFP) and BBa_E0430 (eYFP) into pSB1C3 transformed both ΔEnvZ (JW3367-3) and wt (DH5a) <i>E. coli</i> strains with them. Constructs were validated by sequencing. We submitted the constructs as: <html><a href="https://parts.igem.org/Part:BBa_K1909013">BBa_K1909013</a></html> (Omp/GFP) and <html><a href="https://parts.igem.org/Part:BBa_K1909014">BBa_K1909014</a></html> (Omp/eYFP) to the registry.</p> | ||
| + | |||
| + | <h3>Experiments</h3> | ||
| + | <p> | ||
| + | To determine the noise of the OmpR promoter, we measured the expression of our Fluorescence reporters by flow cytometry, comparing the ΔEnvZ (JW3367-3) to wt (DH5a) <i>E. coli</i> strain. Furthermore, we used JW3367-3 and DH5a strains without our reporter system as negative controls. Cells were incubated in LB medium to OD<sub>600</sub> = 0.2, washed using 100 mM MgCl<sub>2</sub> and diluted 1:100 prior to measuring.</p> | ||
| + | |||
| + | <h3>Results</h3> | ||
| + | <p> | ||
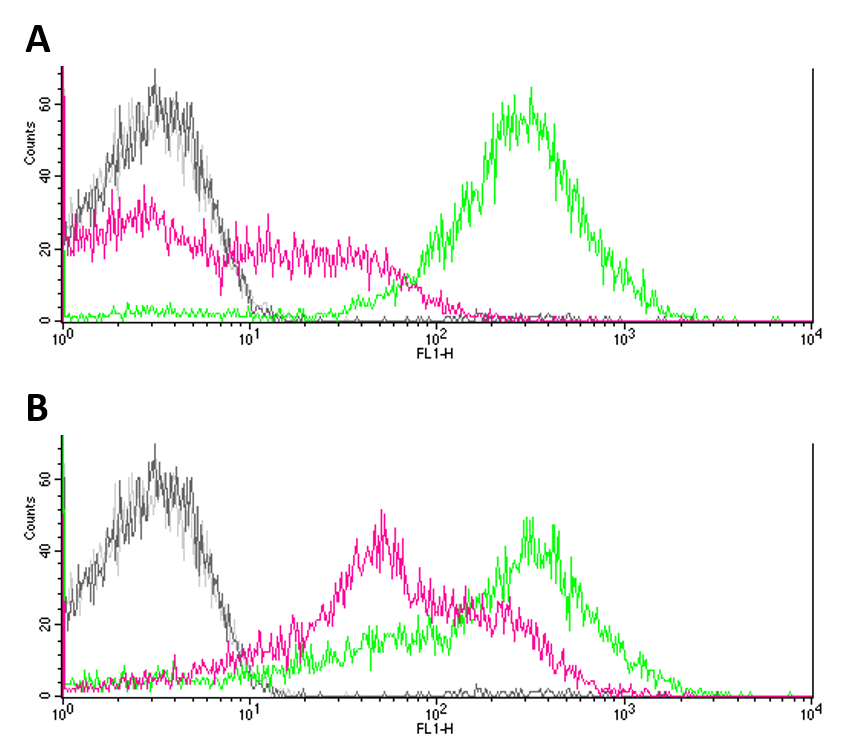
| + | Negative controls (JW3367-3 and DH5a without reporter systems) showed only few to no fluorescence (Fig.1 A, B). GFP expression (Fig. 1 A) proved to be significantly higher in DH5a (green) cells as compared to EnvZ deficient JW3367-3 cells (pink). Although not as pronounced, this difference was also observable for eYFP expression in the respective cell lines (fig. 1 B). Differences in the expression of fluorescence reporters were confirmed by KS tests (GFP: p <= 0.001, eYFP: p <= 0.001). </p> | ||
| + | [[Image:BBa_R0082-FACS_noise.png|500px]] | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
Revision as of 18:55, 20 October 2016
Promoter (OmpR, positive)
Positively regulated, OmpR-controlled promoter. This promoter is taken from the upstream region of ompC. Phosphorylated OmpR binds to the three operator sites and activates transcription.
Usage and Biology
In nature, this promoter is upstream of the ompC porin gene. The regulation of ompC is determined by the EnvZ-OmpR osmosensing machinery. EnvZ phosphorylates OmpR to OmpR-P. At high osmolarity, EnvZ is more active, creating more OmpR-P. OmpR-P then binds to the low-affinity OmpR operator sites upstream of ompC.
Contribution: Hamburg 2016
Group: Hamburg / Authors: Kai Pohl, Daniel Wedemeyer
Summary: We analysed the noise produced by BBa_R0082 by flow cytometry using a ΔEnvZ strain.
In two approaches, we ligated BBa_R0082 (OmpR, positive promoter) with BBa_E0240 (GFP) and BBa_E0430 (eYFP) into pSB1C3 transformed both ΔEnvZ (JW3367-3) and wt (DH5a) E. coli strains with them. Constructs were validated by sequencing. We submitted the constructs as: BBa_K1909013 (Omp/GFP) and BBa_K1909014 (Omp/eYFP) to the registry.
Experiments
To determine the noise of the OmpR promoter, we measured the expression of our Fluorescence reporters by flow cytometry, comparing the ΔEnvZ (JW3367-3) to wt (DH5a) E. coli strain. Furthermore, we used JW3367-3 and DH5a strains without our reporter system as negative controls. Cells were incubated in LB medium to OD600 = 0.2, washed using 100 mM MgCl2 and diluted 1:100 prior to measuring.
Results
Negative controls (JW3367-3 and DH5a without reporter systems) showed only few to no fluorescence (Fig.1 A, B). GFP expression (Fig. 1 A) proved to be significantly higher in DH5a (green) cells as compared to EnvZ deficient JW3367-3 cells (pink). Although not as pronounced, this difference was also observable for eYFP expression in the respective cell lines (fig. 1 B). Differences in the expression of fluorescence reporters were confirmed by KS tests (GFP: p <= 0.001, eYFP: p <= 0.001).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]