Difference between revisions of "Part:pSB1A10"
| Line 6: | Line 6: | ||
[[Image:ScreeningPlasmid1.0.PNG|600px|left|thumb|'''Design of Screening Plasmid 1.0:''' We are using the Pbad arabinose-inducible induction system [1] as a tunable input. GFP is a measure of input and RFP is a measure of output. A Biobricks cloning site enables easy insertion of any Biobricks part. RNase E sites create independence between the mRNA stability of the device being screened and the mRNA stability of the fluorescent proteins. In particular, we suspect mRFP1 contains internal RNaseE cut sites and have added a hairpin 5’ of the coding region to slow degradation by RNase E. [2] | [[Image:ScreeningPlasmid1.0.PNG|600px|left|thumb|'''Design of Screening Plasmid 1.0:''' We are using the Pbad arabinose-inducible induction system [1] as a tunable input. GFP is a measure of input and RFP is a measure of output. A Biobricks cloning site enables easy insertion of any Biobricks part. RNase E sites create independence between the mRNA stability of the device being screened and the mRNA stability of the fluorescent proteins. In particular, we suspect mRFP1 contains internal RNaseE cut sites and have added a hairpin 5’ of the coding region to slow degradation by RNase E. [2] | ||
]] | ]] | ||
| − | + | <br style="clear:both" /> | |
===Usage and Biology=== | ===Usage and Biology=== | ||
Revision as of 09:27, 26 May 2006
Screening Plasmid v1.0
This plasmid can be used to do a rough characterization of the Input/Output curve for a PoPS-based device. Additionally, it can be used for screening of part or device libaries via FACS. More information can be [http://openwetware.org/index.php?title=Endy:Screening_plasmid_1.0 found here.]
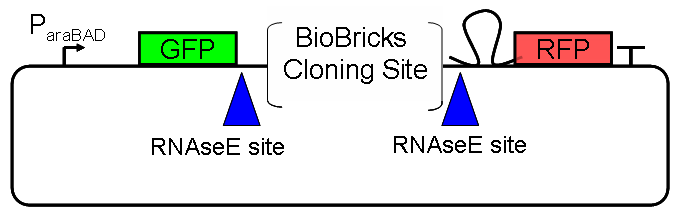
Design of Screening Plasmid 1.0: We are using the Pbad arabinose-inducible induction system [1] as a tunable input. GFP is a measure of input and RFP is a measure of output. A Biobricks cloning site enables easy insertion of any Biobricks part. RNase E sites create independence between the mRNA stability of the device being screened and the mRNA stability of the fluorescent proteins. In particular, we suspect mRFP1 contains internal RNaseE cut sites and have added a hairpin 5’ of the coding region to slow degradation by RNase E. [2]
Usage and Biology
-- Please enter your experience with this part here --
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 5242
Illegal NheI site found at 4443
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NotI site found at 9
Illegal NotI site found at 5248 - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 5242
Illegal BglII site found at 89
Illegal BamHI site found at 4382
Illegal XhoI site found at 67 - 23INCOMPATIBLE WITH RFC[23]Illegal prefix found at 5242
Illegal suffix found at 2 - 25INCOMPATIBLE WITH RFC[25]Illegal prefix found at 5242
Plasmid lacks a suffix.
Illegal XbaI site found at 5257
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal AgeI site found at 674
Illegal AgeI site found at 786
Illegal AgeI site found at 4217 - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal BsaI.rc site found at 2197
Illegal BsaI.rc site found at 5118
Illegal SapI site found at 4199
Functional Parameters
| chassis | -NA- |
| copies | 100-300 |
| mcs | BioBrick |
| origin | pMB1 |
| resistance | A |
References
[1] Khlebnikov et al,Modulation of gene expression from the arabinose-inducible araBAD promoter. J Ind Microbiol Biotechnol. 2002 Jul;29(1):34-7.
[2] Effect of gene location, mRNA secondary structures, and RNase sites on expression of two genes in an engineered operon. Biotechnol Bioeng. 2002 Dec 30;80(7):762-76.
