Difference between revisions of "Part:BBa K4000001"
| Line 4: | Line 4: | ||
| − | <html> | + | <!DOCTYPE html> |
| + | <html lang="en"> | ||
<head> | <head> | ||
| − | <style> | + | <meta charset="UTF-8"> |
| − | + | <meta name="viewport" content="width=device-width, initial-scale=1.0"> | |
| − | + | <title>Contribution by iGEM23_SubCat-China</title> | |
| − | + | <style> | |
| − | </style> | + | .centered { |
| + | display: block; | ||
| + | margin-left: auto; | ||
| + | margin-right: auto; | ||
| + | width: 600px; | ||
| + | } | ||
| + | |||
| + | .centered-text { | ||
| + | text-align: center; | ||
| + | } | ||
| + | </style> | ||
</head> | </head> | ||
<body> | <body> | ||
| − | < | + | <h3>Improvement by Team iGEM23_SubCat-China</h3> |
| + | <h4>BBa _ K4000001 (GA)</h4> | ||
| + | <p>Group: SubCat-China iGEM 2023</p> | ||
| − | < | + | <h4>Summary:</h4> |
| − | < | + | <p>We constructed a new recombinant plasmid BBa _ K4845020( X-2-GA-2 ) based on the old part BBa _ K4000001 (GA)...</p> |
| + | <h4>Construction Design and Engineering Principle</h4> | ||
| + | <p>The problems we are going to solve are... [the rest of your text]...</p> | ||
| − | < | + | <img src="https://static.igem.wiki/teams/4845/wiki/fig-1-overview-of-design.jpg" alt="Figure1: Overview of the methodology of our project design" class="centered"> |
| − | + | <p class="centered-text">Figure1: Overview of the methodology of our project design through synthetic biology (made in Canva.cn)</p> | |
| − | <p> | + | |
| − | < | + | <h4>Characterization/Measurement</h4> |
| − | < | + | <h5>A)Method of Transparent Circle</h5> |
| − | < | + | <p>According to the property of starch... [the rest of your text]...</p> |
| − | + | ||
| − | </ | + | |
| − | < | + | <img src="https://static.igem.wiki/teams/4845/wiki/figure-2-transparent-circle-experiment-for-the-function-testing.png" alt="Figure 2: Transparent circle experiment for the function testing" class="centered"> |
| + | <p class="centered-text">Figure 2: Transparent circle experiment for the function testing...</p> | ||
| − | < | + | <h5>B)Enzyme Activity Detection in Starch Hydrolyzing Capacity</h5> |
| − | <p> | + | <p>To verify the GA and temA activity... [the rest of your text]...</p> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | <img src="https://static.igem.wiki/teams/4845/wiki/figure-3-the-enzyme-activity-of-yeast-1974-ga-tema-and-wild-yeast-1974-under-different-ph-value-at-the-same-temperature.png" alt="Figure 3: The enzyme activity of Yeast 1974-GA-temA and Wild Yeast 1974" class="centered"> | |
| − | + | <p class="centered-text">Figure 3: The enzyme activity of Yeast 1974-GA-temA and Wild Yeast 1974...</p> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | <p class=" | + | |
| − | + | <img src="https://static.igem.wiki/teams/4845/wiki/figure-4-the-enzyme-activity-of-yeast-1974-ga-tema-and-wild-yeast-1974-under-different-temperature-at-the-same-ph-value.png" alt="Figure 4: The enzyme activity of Yeast 1974-GA-temA and Wild Yeast 1974" class="centered"> | |
| − | + | <p class="centered-text">Figure 4: The enzyme activity of Yeast 1974-GA-temA and Wild Yeast 1974...</p> | |
| − | </ | + | |
| − | < | + | <img src="https://static.igem.wiki/teams/4845/wiki/figure-5-the-comparison-of-the-enzyme-activity-curves-of-yeast-1974-ga-tema-under-different-ph-values-and-different-temperature-respectively.png" alt="Figure 5: The comparison of the enzyme activity curves of Yeast 1974-GA-temA" class="centered"> |
| + | <p class="centered-text">Figure 5: The comparison of the enzyme activity curves of Yeast 1974-GA-temA...</p> | ||
| − | < | + | <h5>C)Determination of alcohol production</h5> |
| − | + | <p>To further and directly verify... [the rest of your text]...</p> | |
| − | </ | + | |
| − | <p class=" | + | <img src="https://static.igem.wiki/teams/4845/wiki/figure-6-alcohol-production-of-yeast-1974-and-ga-tema-1974.png" alt="Figure 6: Alcohol production of yeast 1974 and GA-temA-1974" class="centered"> |
| + | <p class="centered-text">Figure 6: Alcohol production of yeast 1974 and GA-temA-1974</p> | ||
| − | < | + | <h4>Reference:</h4> |
| − | + | <ol> | |
| − | + | <li>[2] Xu Shuai, Yang Li, Tan Liping, et al. Enzymatic hydrolysis and application of sweet potato residue [ J ]. Journal of Qilu University of Technology. 2021, 35(03): 28-33. | |
| − | + | [3] Xin Wang, Bei Liao, Zhijun Li, et al. Reducing glucoamylase usage for commercial-scale ethanol production from starch using glucoamylase expressing Saccharomyces cerevisiae[J]. Bioresources and Bioprocessing, 2021, 8(1): 20. | |
| − | </ | + | [4] Rosemary A. Cripwell, Lorenzo Favaro, Marinda Viljoen-Bloom, et al. Consolidated bioprocessing of raw starch to ethanol by Saccharomyces cerevisiae: Achievements and challenges[J]. Biotechnology Advances, 2020, 42: 107579. |
| − | + | [5] LM de Moraes, S Astolfi-Filho, SG Oliver. Development of yeast strains for the efficient utilisation of starch: evaluation of constructs that express alpha-amylase and glucoamylase separately or as bifunctional fusion proteins[J]. Applied microbiology and biotechnology, 1995, 43(6): 1067-76.</li> | |
| − | < | + | </ol> |
| − | + | ||
| − | < | + | |
| − | + | ||
| − | + | ||
| − | + | ||
</body> | </body> | ||
</html> | </html> | ||
| + | |||
Revision as of 10:26, 9 October 2023
GA
<!DOCTYPE html>
Improvement by Team iGEM23_SubCat-China
BBa _ K4000001 (GA)
Group: SubCat-China iGEM 2023
Summary:
We constructed a new recombinant plasmid BBa _ K4845020( X-2-GA-2 ) based on the old part BBa _ K4000001 (GA)...
Construction Design and Engineering Principle
The problems we are going to solve are... [the rest of your text]...

Figure1: Overview of the methodology of our project design through synthetic biology (made in Canva.cn)
Characterization/Measurement
A)Method of Transparent Circle
According to the property of starch... [the rest of your text]...

Figure 2: Transparent circle experiment for the function testing...
B)Enzyme Activity Detection in Starch Hydrolyzing Capacity
To verify the GA and temA activity... [the rest of your text]...

Figure 3: The enzyme activity of Yeast 1974-GA-temA and Wild Yeast 1974...

Figure 4: The enzyme activity of Yeast 1974-GA-temA and Wild Yeast 1974...

Figure 5: The comparison of the enzyme activity curves of Yeast 1974-GA-temA...
C)Determination of alcohol production
To further and directly verify... [the rest of your text]...

Figure 6: Alcohol production of yeast 1974 and GA-temA-1974
Reference:
- [2] Xu Shuai, Yang Li, Tan Liping, et al. Enzymatic hydrolysis and application of sweet potato residue [ J ]. Journal of Qilu University of Technology. 2021, 35(03): 28-33. [3] Xin Wang, Bei Liao, Zhijun Li, et al. Reducing glucoamylase usage for commercial-scale ethanol production from starch using glucoamylase expressing Saccharomyces cerevisiae[J]. Bioresources and Bioprocessing, 2021, 8(1): 20. [4] Rosemary A. Cripwell, Lorenzo Favaro, Marinda Viljoen-Bloom, et al. Consolidated bioprocessing of raw starch to ethanol by Saccharomyces cerevisiae: Achievements and challenges[J]. Biotechnology Advances, 2020, 42: 107579. [5] LM de Moraes, S Astolfi-Filho, SG Oliver. Development of yeast strains for the efficient utilisation of starch: evaluation of constructs that express alpha-amylase and glucoamylase separately or as bifunctional fusion proteins[J]. Applied microbiology and biotechnology, 1995, 43(6): 1067-76.
Profile
Name: GA
Base Pairs: 1567bp
Origin: Saccharomycopsis fibuligera, synthesis
Properties: An enzyme that can easily break down starches into glucose
Usage and Biology
Glucoamylase is an enzyme that can be obtained from the yeast or fungi in the Aspergillus genus such as Aspergillus niger. The enzyme decomposes starch molecules in the human body into the useful energy compound of glucose. This is accomplished by removing the alpha-1 and 4-glycosidic linkages from the non-reducing end of the starch molecule. These molecules are more commonly referred to as polysaccharides and are frequently either amylase- or amylopectin-based.
Experimental approach
1.GA plasmids transformation
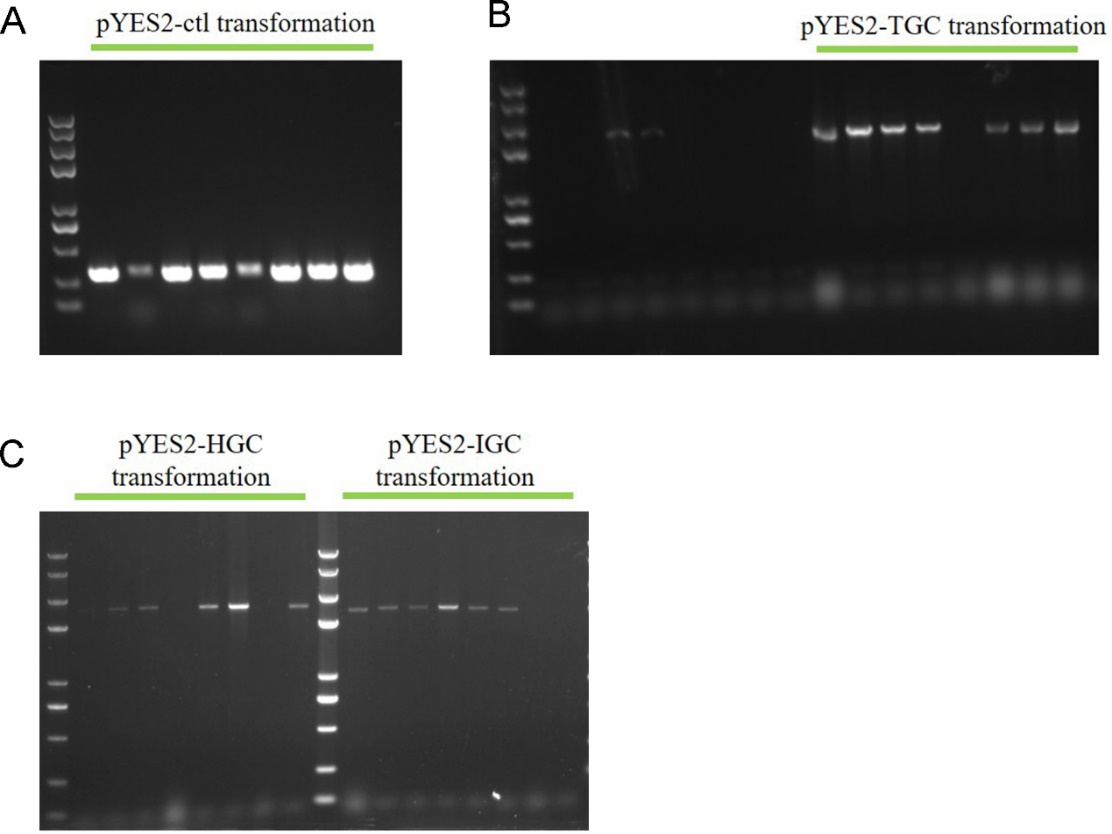
GA-expressing plasmids transformation and positive S. cerevisiae transformants selection using 50 μg/mL (Figure 2A) and 350 μg/mL (Figure 2B) hygromycin. Top left, pYES2-ctl. Top right, pYES2-TGC. Bottom left, pYES2-HGC. Bottom right, pYES2-IGC.
2. Colony PCR to identify the correct transpormed S. cerevisiae
The colonies which grew on the high concentration hygromycin plates were subjected to colony PCR to verify the plasmids transformation again. From Fig. 3 we can see that positive bands implied the plasmids transformation successfully.
Proof of function
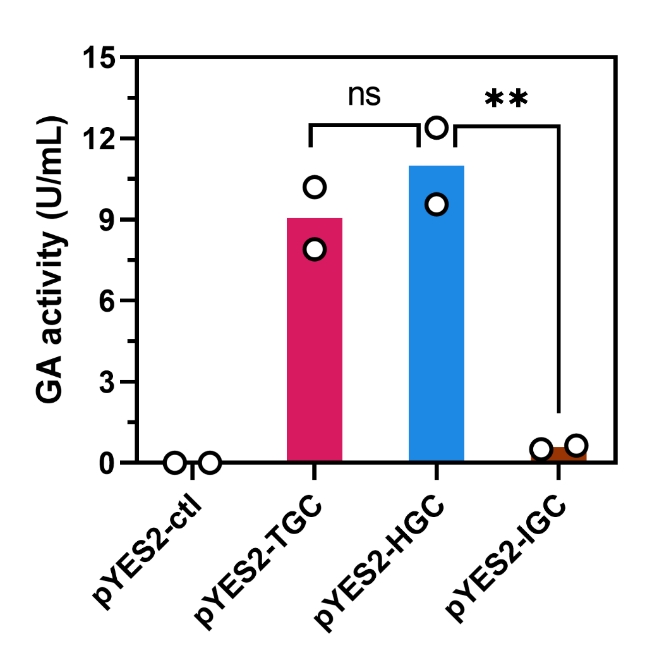
1. Glucoamylase activity assay
S. cerevisiae strains harboring various plasmids glucoamylase activity determination. **Statistical significance between indicated strains by Student’s t test, p < 0.01. ns, not significant. Data represent the means of two independent colonies.
2. Fermentation test
To verify the GA secretion capacity of the GA-expressing S. cerevisiae strains, we measured the glucose concentration inside the cell during the corn starch fermentation process. At the initial stage (0 h), when the GA was added during the “starch-to-glucose” process, higher contents of glucose were detected than the process without GA addition.
References
1. Görgens J F, Bressler D C, van Rensburg E. Engineering Saccharomyces cerevisiae for direct conversion of raw, uncooked or granular starch to ethanol[J]. Critical reviews in biotechnology, 2015, 35(3): 369-391.
2. Van Zyl W H, Bloom M, Viktor M J. Engineering yeasts for raw starch conversion[J]. Applied microbiology and biotechnology, 2012, 95(6): 1377-1388.
3. Maury J, Kannan S, Jensen N B, et al. Glucose-dependent promoters for dynamic regulation of metabolic pathways[J]. Frontiers in bioengineering and biotechnology, 2018, 6: 63.
4. Weber E, Engler C, Gruetzner R, et al. A modular cloning system for standardized assembly of multigene constructs[J]. PloS one, 2011, 6(2): e16765.
5. Pollak B, Cerda A, Delmans M, et al. Loop assembly: a simple and open system for recursive fabrication of DNA circuits[J]. New Phytologist, 2019, 222(1): 628-640.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 131
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 92
Illegal AgeI site found at 1463 - 1000COMPATIBLE WITH RFC[1000]