Difference between revisions of "Part:BBa K4182011"
| Line 34: | Line 34: | ||
==Usage&Biology== | ==Usage&Biology== | ||
| − | + | Biosafety is an important consideration when designing engineered bacteria, and MazF has been commonly used in previously work. Here we develop a novel lysis gene (Gene ID: IF654_RS00240) for the design of controlled suicide circuits for engineering efficiency and biosafety reasons, also can be considered as an alternative of suicide protein MazF. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | Biosafety is an important consideration when designing engineered bacteria | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
[[File:XJTU-p5-1.png|500px]] | [[File:XJTU-p5-1.png|500px]] | ||
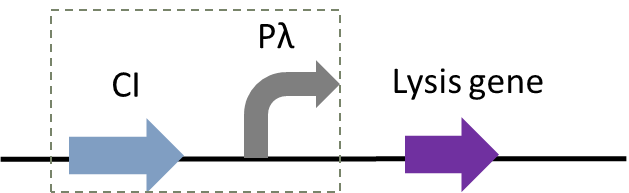
| − | Figure 1: | + | Figure 1: The temperature-controlled suicide circuit with a novel lysis gene |
[[File:XJTU-p5-2.png|500px]] | [[File:XJTU-p5-2.png|500px]] | ||
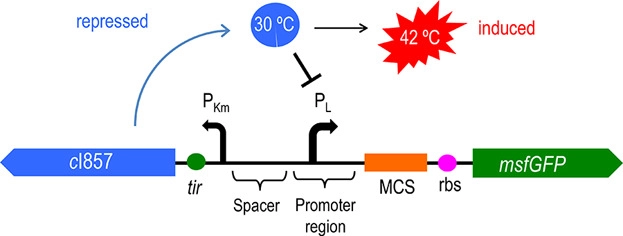
| − | Figure 2 | + | Figure 2: The principle of temperature-controlled suicide circuit |
| − | + | Figure 1 and Figure 2 shows the principle of our temperature-controlled suicide circuit. When bacteria grows at a low temperature(30℃), the CI857 protein binds to the Pλ promoter, and downstream lysis gene are unable to be expressed, allowing the cell growth. While at 42℃, the CI protein will be degraded and lead to the expression of lysis gene and eventually cell death and release of the product. | |
| − | ==Codon | + | ===Codon optimization of lysis gene=== |
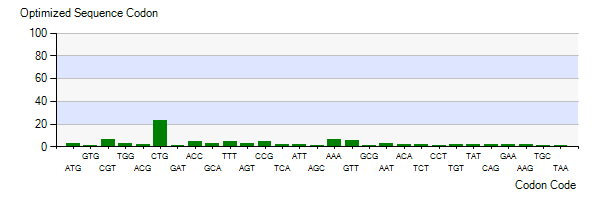
| − | The | + | The lysis gene is from Enterobacteria phage KleenX174. To better express the lysis gene in engineered bacteria, the codon optimization of the lysis gene was conducted according to the codon preference of Escherichia coli. Figure 3 shows the number of codons we optimized to make our codons more in line with Escherichia coli preference. The modified lysis gene is shown in BBa_K4182007. |
[[File:XJTU-p5-3.png|500px]] | [[File:XJTU-p5-3.png|500px]] | ||
| − | Figure 3: | + | Figure 3: Codon optimization of lysis gene |
| − | == | + | ===Construction and optimization of suicide circuit=== |
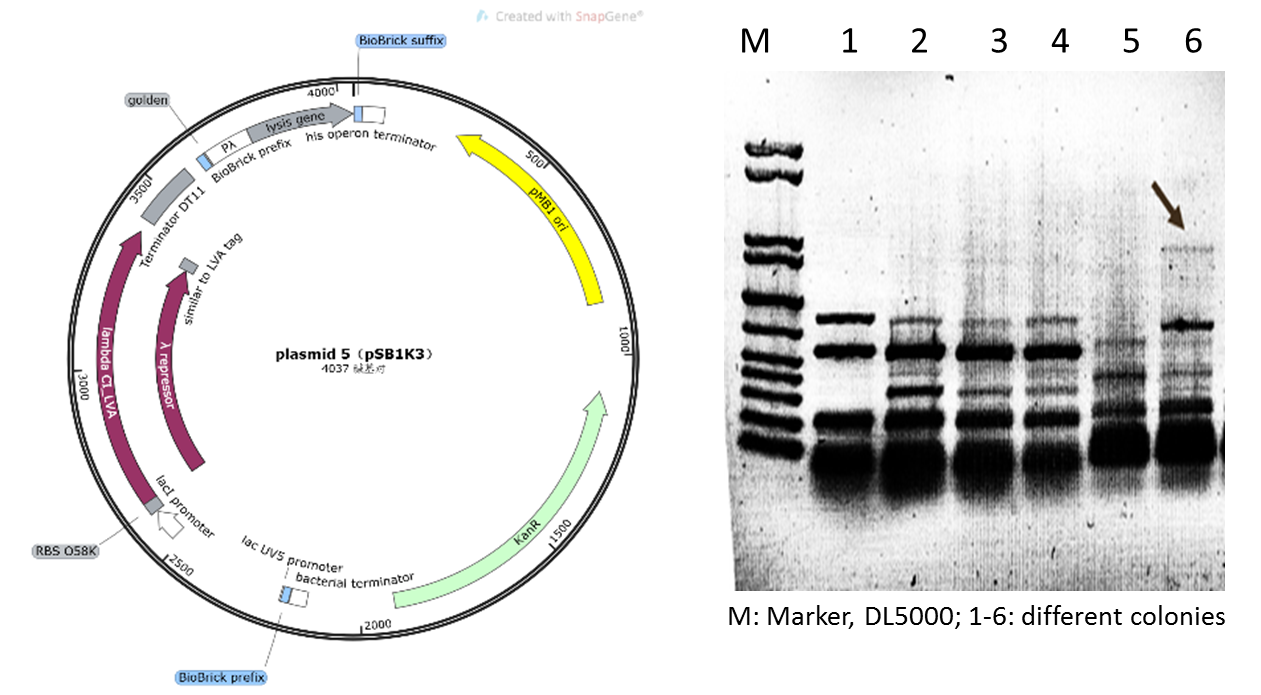
| − | Initially | + | Initially, we planned to construct our suicide circuit (Plasmid 5) using vector backbone pSB1K3 by Golden Gate assembly, and we can obtain several clones. However, it was found that after colony PCR verification, only weak target bands could be observed (Figure 4), and the plasmids extracted from the recombinant DH5α cells was at very low concentration, and sequencing could not be completed. |
[[File:XJTU-p5-10.png|500px]] | [[File:XJTU-p5-10.png|500px]] | ||
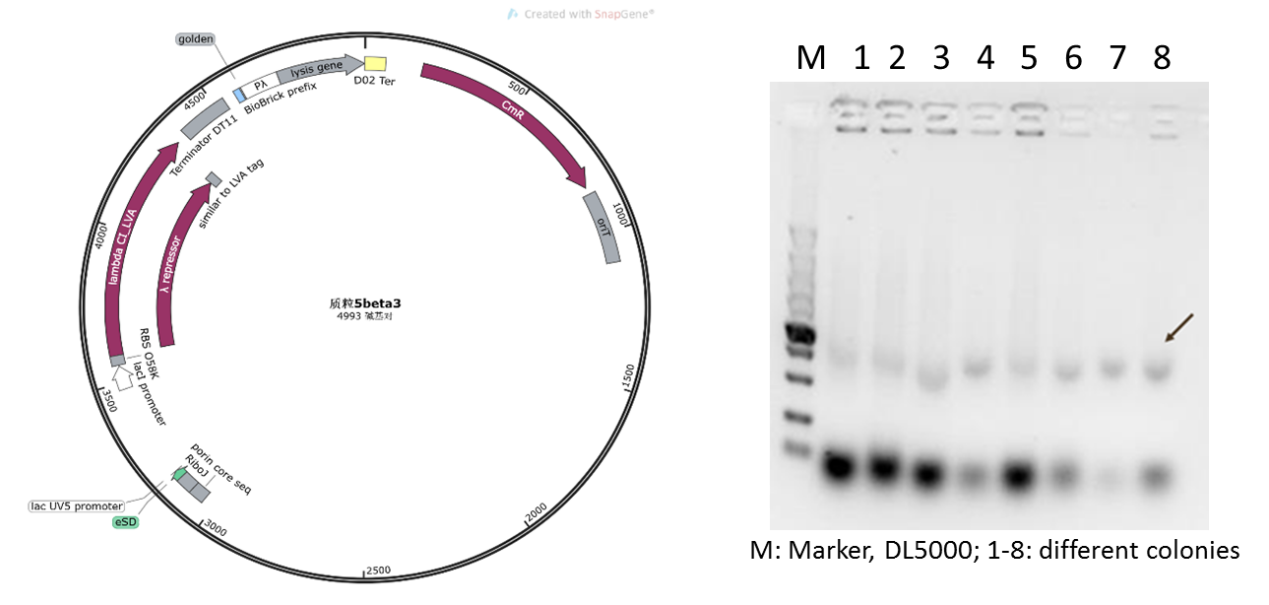
| − | Figure 4: Plasmid 5 map based on pSB1K3 | + | Figure 4: Plasmid 5 map based on pSB1K3 and its verification (he target band is approximately 1600bp) |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | We supposed that due to the low copy number of pSB1K3 vector, it is difficult to extract sufficient plasmid from the engineered bacteria for further validation. Therefore, we replaced the backbone to pSEVA341, a higher-copy-number vector and re-constructed the plasmid (Figure 5). As shown in Figure 6, the obvious target bands were observed, and the plasmid correctness was further confirmed by sequencing. | |
| − | + | [[File:XJTU-p5-11.png|500px]] | |
| − | + | Figure 5: the new plasmid 5 with pSEVA341 backbone and its verification | |
| − | === | + | ===The verification of heat triggered cell lysis and suicide=== |
| + | The engineered cell harboring plasmid 5 and blank vector respectively, were culture at 30℃ overnight, and then the temperature was shift to 42℃. The OD600 of each group was detected every 1 h, and the growth curve of these strains were determined as follows. | ||
| + | The results clearly demonstrated the cell growth was significantly inhibited after heat at 42℃ compared to the strain without lysis gene. And about 9% of whole cell population was retained after heat. Compared to the commonly used suicide protein MazF (BBa_K302033), the lysis protein in our study is also efficient but shorter and easy to be manipulated, which can be used as an alternative and update for MazF. | ||
| − | |||
| − | [[File:XJTU-p5- | + | [[File:XJTU-p5-12.png|500px]] |
| − | Figure | + | Figure 6: The cell growth of strains with suicide plasmid 5 and blank vector |
==References== | ==References== | ||
Revision as of 18:45, 13 October 2022
Temperature regulated suicide circuit
In addition to population-responsive suicide mechanisms, suicide systems with other regulatory modes can also be designed through synthetic biology. Here, we designed a temperature-responsive cleavage system to achieve temperature-controlled cleavage, that is, cleavage of thermoregulated lysis genes (Gene ID: IF654_RS00240)
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal prefix found in sequence at 1018
Illegal suffix found in sequence at 1446 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 1018
Illegal SpeI site found at 1447
Illegal PstI site found at 1461
Illegal NotI site found at 1024
Illegal NotI site found at 1454 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 1018
Illegal XhoI site found at 2478
Illegal XhoI site found at 3504 - 23INCOMPATIBLE WITH RFC[23]Illegal prefix found in sequence at 1018
Illegal suffix found in sequence at 1447 - 25INCOMPATIBLE WITH RFC[25]Illegal prefix found in sequence at 1018
Illegal XbaI site found at 1033
Illegal SpeI site found at 1447
Illegal PstI site found at 1461 - 1000COMPATIBLE WITH RFC[1000]
Profile
Base Pairs
3733
Design Notes
This gene has been optimized for E. coli
Source
Escherichia phage phiX174(from https://www.ncbi.nlm.nih.gov/gene/2546400)
Usage&Biology
Biosafety is an important consideration when designing engineered bacteria, and MazF has been commonly used in previously work. Here we develop a novel lysis gene (Gene ID: IF654_RS00240) for the design of controlled suicide circuits for engineering efficiency and biosafety reasons, also can be considered as an alternative of suicide protein MazF.
Figure 1: The temperature-controlled suicide circuit with a novel lysis gene
Figure 2: The principle of temperature-controlled suicide circuit
Figure 1 and Figure 2 shows the principle of our temperature-controlled suicide circuit. When bacteria grows at a low temperature(30℃), the CI857 protein binds to the Pλ promoter, and downstream lysis gene are unable to be expressed, allowing the cell growth. While at 42℃, the CI protein will be degraded and lead to the expression of lysis gene and eventually cell death and release of the product.
Codon optimization of lysis gene
The lysis gene is from Enterobacteria phage KleenX174. To better express the lysis gene in engineered bacteria, the codon optimization of the lysis gene was conducted according to the codon preference of Escherichia coli. Figure 3 shows the number of codons we optimized to make our codons more in line with Escherichia coli preference. The modified lysis gene is shown in BBa_K4182007.
Figure 3: Codon optimization of lysis gene
Construction and optimization of suicide circuit
Initially, we planned to construct our suicide circuit (Plasmid 5) using vector backbone pSB1K3 by Golden Gate assembly, and we can obtain several clones. However, it was found that after colony PCR verification, only weak target bands could be observed (Figure 4), and the plasmids extracted from the recombinant DH5α cells was at very low concentration, and sequencing could not be completed.
Figure 4: Plasmid 5 map based on pSB1K3 and its verification (he target band is approximately 1600bp)
We supposed that due to the low copy number of pSB1K3 vector, it is difficult to extract sufficient plasmid from the engineered bacteria for further validation. Therefore, we replaced the backbone to pSEVA341, a higher-copy-number vector and re-constructed the plasmid (Figure 5). As shown in Figure 6, the obvious target bands were observed, and the plasmid correctness was further confirmed by sequencing.
Figure 5: the new plasmid 5 with pSEVA341 backbone and its verification
The verification of heat triggered cell lysis and suicide
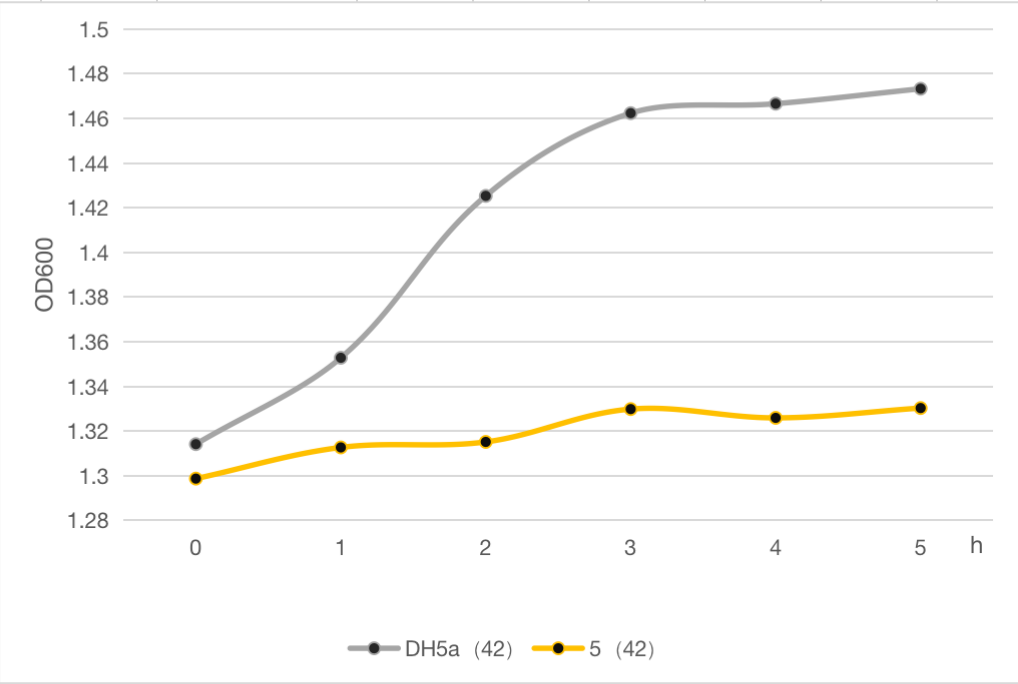
The engineered cell harboring plasmid 5 and blank vector respectively, were culture at 30℃ overnight, and then the temperature was shift to 42℃. The OD600 of each group was detected every 1 h, and the growth curve of these strains were determined as follows. The results clearly demonstrated the cell growth was significantly inhibited after heat at 42℃ compared to the strain without lysis gene. And about 9% of whole cell population was retained after heat. Compared to the commonly used suicide protein MazF (BBa_K302033), the lysis protein in our study is also efficient but shorter and easy to be manipulated, which can be used as an alternative and update for MazF.
Figure 6: The cell growth of strains with suicide plasmid 5 and blank vector
References
1. Din, M.O., et al., Synchronized cycles of bacterial lysis for in vivo delivery. Nature, 2016. 536(7614): p. 81-85.
2. Saeidi, N., et al., Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol Syst Biol, 2011. 7: p. 521.
3. Restrepo-Pineda, S., et al., Thermoinducible expression system for producing recombinant proteins in Escherichia coli: advances and insights. FEMS Microbiol Rev, 2021. 45(6).
4. Aparicio, T., V. de Lorenzo, and E. Martínez-García, Improved Thermotolerance of Genome-Reduced Pseudomonas putida EM42 Enables Effective Functioning of the PL/cI857 System. Biotechnology Journal, 2019. 14(1): p. 1800483.