Difference between revisions of "Part:BBa K4274010"
| Line 9: | Line 9: | ||
<br>In this case, DNA sequence of degQ gene under the regulation of p43 promoter was knocked-in the region of the natural sfp gene of <i>B. subtilis</i>. It was used in the composite part PvanP*-sfp_target-sfp_HA_US-p43-K4274013-sfp-K4274014-degQ-sfp_HA_DS (Part: BBa_K4274035) to realize fengycins’ production in <i>B. subtilis</i>. This can be used for other teams working on fengycin production or research on the antibiotic properties of cyclolipopeptides. | <br>In this case, DNA sequence of degQ gene under the regulation of p43 promoter was knocked-in the region of the natural sfp gene of <i>B. subtilis</i>. It was used in the composite part PvanP*-sfp_target-sfp_HA_US-p43-K4274013-sfp-K4274014-degQ-sfp_HA_DS (Part: BBa_K4274035) to realize fengycins’ production in <i>B. subtilis</i>. This can be used for other teams working on fengycin production or research on the antibiotic properties of cyclolipopeptides. | ||
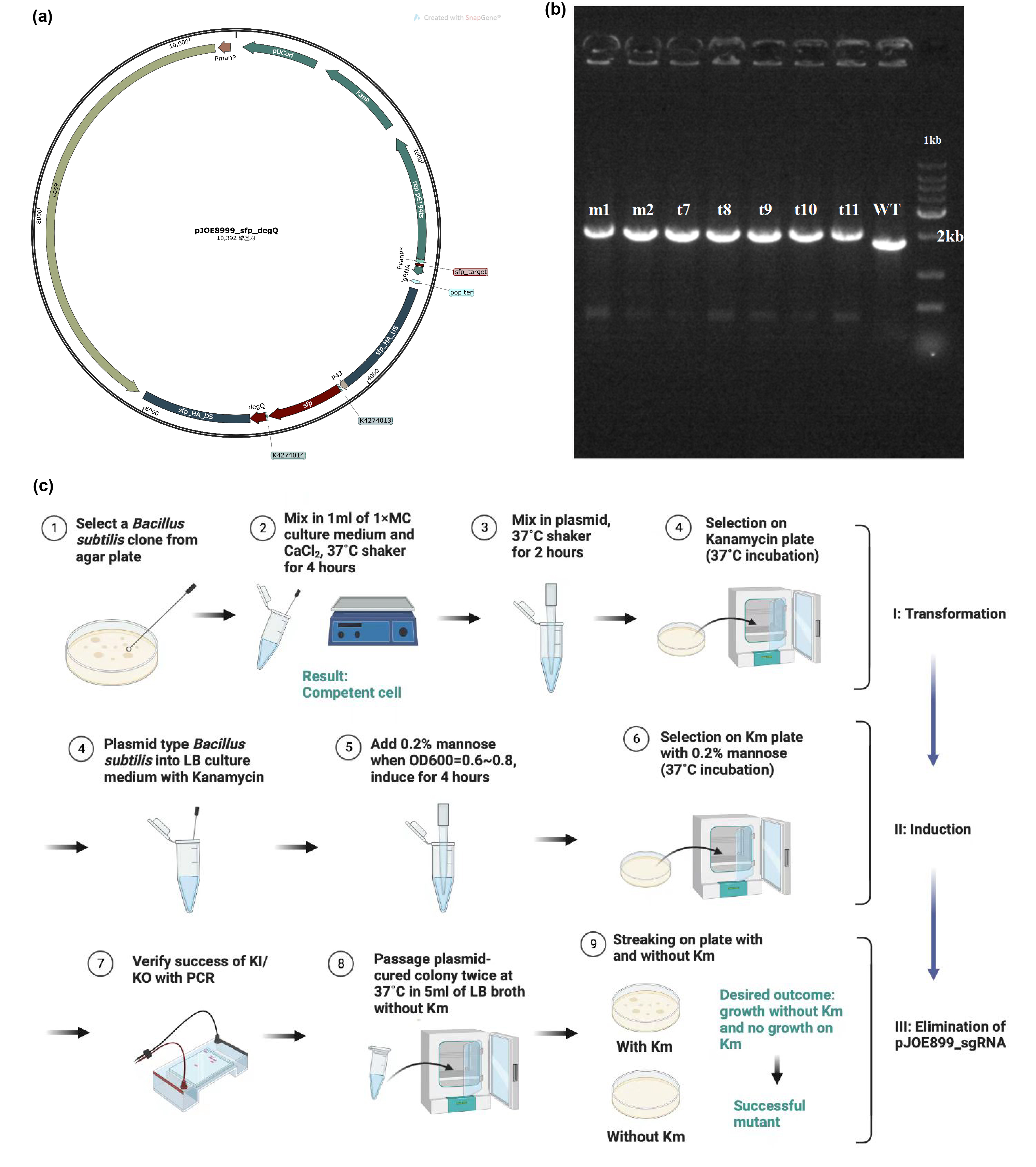
| + | [[Image:Parts-keystone-fengycin1.jpeg|thumbnail|750px|center|'''Figure 1:''' | ||
| + | Edting the genome of <i>Bacillus subtilis</i> 168 to enable it to produce fengycins. (a) Plasmid design for knocking out the invalid sfp gene of <i>Bacillus subtilis</i> 168 and knocking in the sfp gene from B. amyloliquefaciens FZB42 and degQ gene from <i>Bacillus subtilis</i> 168. (b) Electrophoresis results show that gene editing is successful. (c) Protocol about transformation, induction and elimination of pJOE8999 plasmid in <i>Bacillus subtilis</i> 168. ]] | ||
| + | |||
| + | Successful knock-in of the degQ and sfp gene yields sufficient fengycin production in the modified <i>B. subtilis</i>. | ||
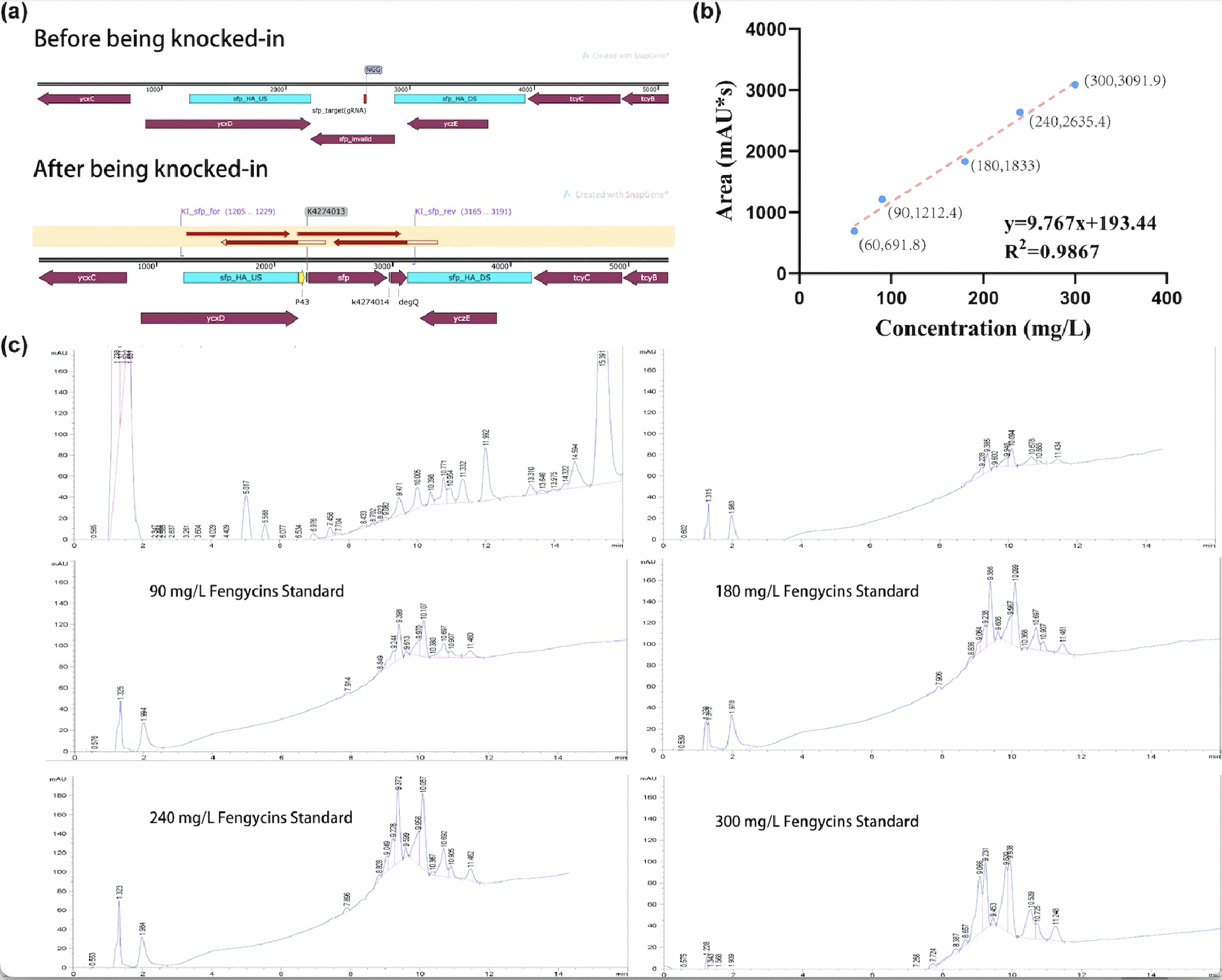
| + | [[Image:Parts-keystone-fengycin2-1.jpeg|750px|thumbnail|center|'''Figure 2:''' | ||
| + | Quantification analysis of fengycins production. (a) The sequencing result of engineering <i>Bacillus subtilis</i> 168 strain for production. (b) The standard curve of peak area obtained by HPLC (y) and its corresponding fengycins' concentration (x). (c) HPLC results of sample and various concentrations of fengycins standard. ]] | ||
==Source== | ==Source== | ||
<i>Bacillus subtilis</i> 168 | <i>Bacillus subtilis</i> 168 | ||
Revision as of 12:03, 12 October 2022
degQ
degQ gene is pleiotropic regulator gene encoding a 46 amino acid polypeptide in Bacillus subtilis 168. It can regulate the expression of a variety of exocrine enzymes and the production of antibacterial substances. Previous studies have shown that the importance of degQ gene in fengycin‘s production. Therefore, degQ gene was knocked-in Bacillus subtilis 168 together with sfp gene from Bacillus amyloliquefaciens FZB42 (Part: BBa_K4274009).
Usage and Biology
degQ gene (Gene ID: 936547) is pleiotropic regulator gene encoding a 46 amino acid polypeptide in Bacillus subtilis 168. It can regulate the expression of a variety of exocrine enzymes and the production of antibacterial substances. It is reported that increased expression of degQ in B. subtilis 168 results in a 7-10 fold increase in antibiotic production. It is because a T base at position -10 in the promoter region of degQ is mutated into a C, resulting in the inability to express degQ.
In this case, DNA sequence of degQ gene under the regulation of p43 promoter was knocked-in the region of the natural sfp gene of B. subtilis. It was used in the composite part PvanP*-sfp_target-sfp_HA_US-p43-K4274013-sfp-K4274014-degQ-sfp_HA_DS (Part: BBa_K4274035) to realize fengycins’ production in B. subtilis. This can be used for other teams working on fengycin production or research on the antibiotic properties of cyclolipopeptides.
Successful knock-in of the degQ and sfp gene yields sufficient fengycin production in the modified B. subtilis.
Source
Bacillus subtilis 168
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
[1]Tsuge K., Ano T., Hirai M., et al. The Genes degQ, pps, and Ipa-8(sfp) Are Responsible for Conversion of Bacillus subtilis 168 to Plipastin Production. Antimicrobial Agents and Chemo. 43(9), 2183-2192 (1999). https://doi.org/10.1128/AAC.43.9.2183.
[2]Gao L., Han J., Liu H., et al. Plipastin and surfactin coproduction by Bacillus subtilis pB2-L and their effects on microorganisms. Antonie van Leeuwenhoek. 110 (3), 1007-1018 (2017). https://doi.org/10.1007/s10482-017-0874-y
[3]Yin Y., Tan W., Wen J., et al. Increasing fengycin production by strengthening the fatty acid synthesis pathway and optimizing fermentation conditions. Biochem. Engineering Journal. 177 (21), 1082 (2021). https://doi.org/10.1016/j.bej.2021.108235
