Difference between revisions of "Part:BBa K4365022"
Svetlana ia (Talk | contribs) |
Svetlana ia (Talk | contribs) |
||
| Line 7: | Line 7: | ||
{| class="wikitable" style="margin-left: auto; margin-right: auto; border: none; width:600px;" | {| class="wikitable" style="margin-left: auto; margin-right: auto; border: none; width:600px;" | ||
| − | |+ Collection of | + | |+ Collection of shuttle vector backbones optimized for ''Saccharomyces cerevisiae'' |
|- | |- | ||
! Backbone ID | ! Backbone ID | ||
Revision as of 12:44, 11 October 2022
pSB1KY-URA3
backbone for shuttle vector. It is compatible with RFC1000 standart.
| Backbone ID | Selection marker | Marker seuqnce source |
|---|---|---|
| BBa_K4365022 | URA3 | [http://freegenes.github.io/genes/BBF10K_000474.html Open Yeast Collection ScURA3-marker] |
| BBa_K4365023 | TRP1 | [http://freegenes.github.io/genes/BBF10K_000473.html Open Yeast Collection ScTRP1-marker] |
| BBa_K4365024 | HIS3 | [http://freegenes.github.io/genes/BBF10K_000468.html Open Yeast Collection ScHIS3-marker] |
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 4632
Illegal XbaI site found at 473
Illegal SpeI site found at 1481
Illegal PstI site found at 12 - 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 4632
Illegal SpeI site found at 1481
Illegal PstI site found at 12
Illegal NotI site found at 2554 - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 4632
Illegal BamHI site found at 108
Illegal XhoI site found at 3482
Illegal XhoI site found at 4508 - 23INCOMPATIBLE WITH RFC[23]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 4632
Illegal XbaI site found at 473
Illegal SpeI site found at 1481
Illegal PstI site found at 12 - 25INCOMPATIBLE WITH RFC[25]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 4632
Illegal XbaI site found at 473
Illegal SpeI site found at 1481
Illegal PstI site found at 12 - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal SapI site found at 4638
Illegal SapI.rc site found at 5
Usage and Biology
Advantages of pSB1KY shuttle vectors
The addition of a yeast origin of replication and an auxotrophy cassette introduced a new function in the original pSB1K plasmid that allows it not only to be expressed in Escherichia coli cells but also in eukaryotic cells of Saccharomyces cerevisiae. We named the shuttle plasmid originating from our genetic work on the pSB1K plasmid pSB1KY (Y stands for yeast).
The origin of replication that we have introduced is a multicopy yeast origin of replication - the 2µOri - and it allows multiple copies to be maintained in yeast cells for increased protein production.
The combination of several auxotrophy cassettes originating from the Open Yeast Collection has allowed us to compile a series of pSB1KY plasmids. The availability of the various auxotrophy cassettes in this plasmid series enhances the accessibility and versatility of the pSB1KY plasmid since users can now choose the pSB1KY that is most suitable for the yeast strain at hand in their laboratory.
These genetic improvements allowed us to take advantage of the RFC1000 assembly standard. Thanks to it we were able to efficiently clone and subsequently screen a library of [signal peptides](https://www.notion.so/5d9471157dd848fc8a210794cf1d6ed0) in *S. cerevisiae.*
The series of pSB1KY plasmids we have created furthermore complements the Open Yeast Collection. Not only is it compatible with its genetic parts but it can also be assembled without the need to put together the whole plasmid from scratch, as is the case for the assembly system on which the Open Yeast Collection is based. This means that the users of the Open Yeast Collection can choose three plasmids out of the collection - a promoter, a CDS, and a terminator - and can assemble them directly into the pSB1KY that is compatible with their lab’s auxotrophic yeast strain.
The pSB1KY plasmids have shown to have great versatility and have found their first users in our partners from WWU_Münster.
pSB1KY-URA3 assembly
The pSB1K plasmid was adapted into shuttle plasmids for the iGEM RFC1000 assembly in S. cerevisiae. We assembled the shuttle pSB1KY shuttle vector (Y stands for yeast).
Inserts carrying genetic elements for the maintenance, selection, and replication of the plasmids in yeast were introduced into the pSB1K. These inserts were a multi-copy 2μ Ori sequence and one of the four of the auxotrophy markers from the Yeast Collection (ScURA3-marker, ScTRP1-marker, ScHIS3-marker, ScLEU2-marker).
Three restriction enzymes were selected from the list of non-cutters in the selection markers, in the backbones, and the 2μ Ori: BamHI, SpeI, and NotI.
These restriction sites were added via primers in a PCR reaction using the plasmids from the Open Yeast Collection as templates. We decided to create a level 1 shuttle plasmid with the *URA3* cassette first and use it then as a base for the cloning of other auxotrophy markers (HIS3, TRP1, LEU2).
The pSB1K4 was linearized by PCR with the primers iGEM level1 BamHI and iGEM level1 NotI (Table 2, Figure 2).
Characterization of the pSB1KY in comparison to the p426 shuttle vector
To understand whether the new functionalities introduced into the pSB1K were suitable for subsequent experiments, we compared the growth curves of the pSB1KY-transformed yeast cells and p426-transformed ones expressing turboRFP. Two main cultures were adjusted to 1 OD in 20 ml of MV medium in flasks and, after 12 hr, their growth rates were monitored using a spectrophotometer.
From the plots of the growth rates (Figure 8) we were able to observe that the new pSB1KY performs just as well as the widely used p426 shuttle plasmid [1].
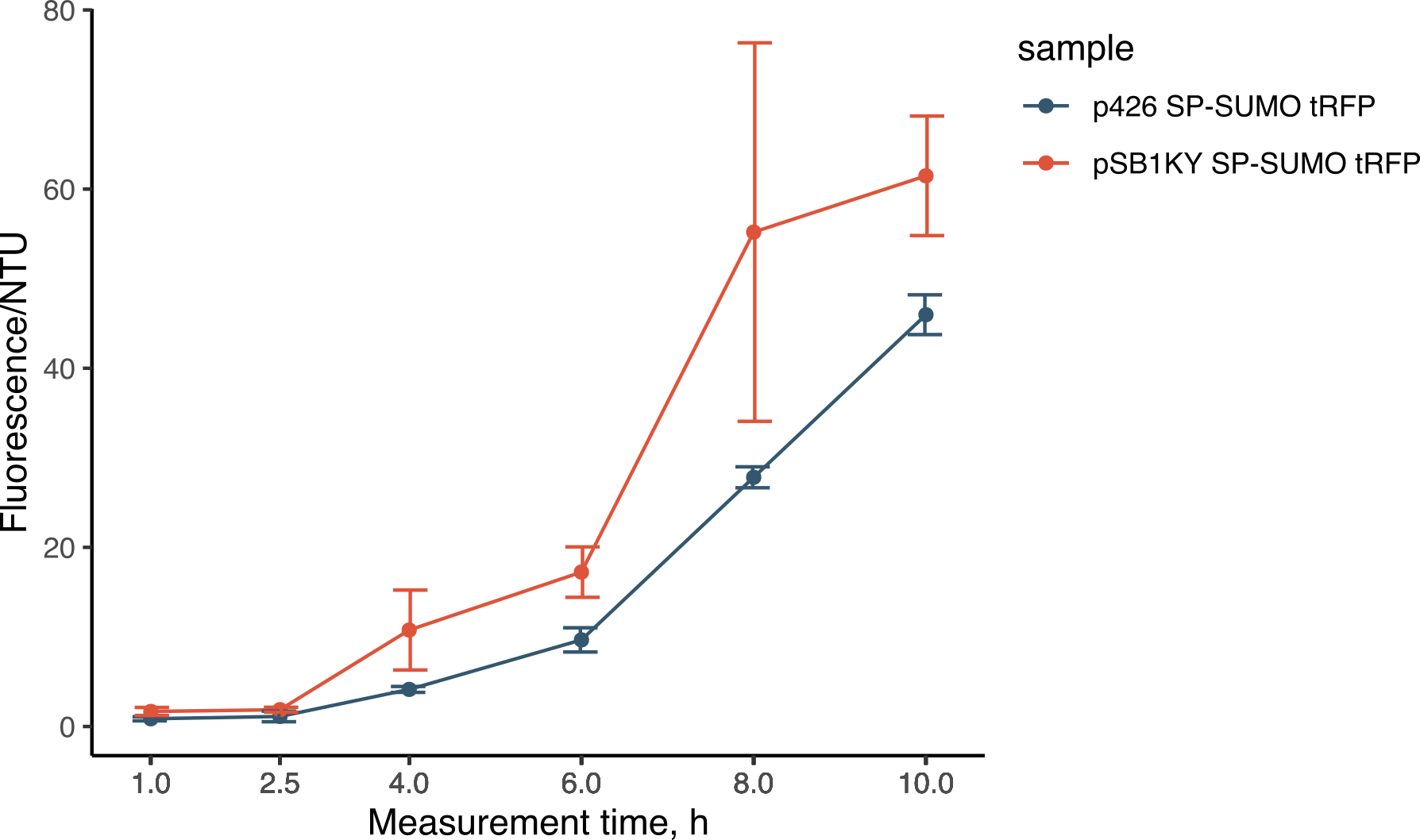
During the experiments for the creation of the SP-SUMO tag for protein production in yeast (read more about it here), we were once again able to compare the two plasmids. Both the p426 and the new pSB1KY were used to express the SP-SUMO device aminoterminally fused to the turboRFP fluorescent protein. This construct allowed the protein to be secreted in the medium and we were able to measure the development of fluorescence. The fluorescence intensity in the medium normalized over growth (expressed in nephelometric turbidity units) (Figure 9) showed that the pSB1KY is able to produce and secrete larger quantities of turboRFP. This was observed in two other experiments (data not shown) and seems to indicate that the pSB1KY allows for higher protein production.