Difference between revisions of "Part:BBa K2981006"
Ahmed Mattar (Talk | contribs) |
Ahmed Mattar (Talk | contribs) (→Characterization by mathematical modeling) |
||
| Line 5: | Line 5: | ||
The part is a signal peptide from actinomycetes, and E.coli can also be secreted normally. The signal peptide efficiently enabled extracellular protein secretion and also contributed to the active expression of the intracellular recombinant proteins. | The part is a signal peptide from actinomycetes, and E.coli can also be secreted normally. The signal peptide efficiently enabled extracellular protein secretion and also contributed to the active expression of the intracellular recombinant proteins. | ||
| − | ==Characterization by mathematical modeling== | + | ==Characterization by mathematical modeling by AFCM-Egypt 2022 team== |
We are using mathematical modeling to detect the increased level of phenylalanine (phe) in phenylketonuria patients in our diagnostic platform. It depends on a whole cell-based biosensor through a cascade of reactions to finally end by formation of β-galactosidase that turns the color into blue once bound to its substrate (X-gal) as mentioned in figure (4) and graph (1). | We are using mathematical modeling to detect the increased level of phenylalanine (phe) in phenylketonuria patients in our diagnostic platform. It depends on a whole cell-based biosensor through a cascade of reactions to finally end by formation of β-galactosidase that turns the color into blue once bound to its substrate (X-gal) as mentioned in figure (4) and graph (1). | ||
Revision as of 13:45, 9 October 2022
Kp-sp, a secreted peptide
The part is a signal peptide from actinomycetes, and E.coli can also be secreted normally. The signal peptide efficiently enabled extracellular protein secretion and also contributed to the active expression of the intracellular recombinant proteins.
Characterization by mathematical modeling by AFCM-Egypt 2022 team
We are using mathematical modeling to detect the increased level of phenylalanine (phe) in phenylketonuria patients in our diagnostic platform. It depends on a whole cell-based biosensor through a cascade of reactions to finally end by formation of β-galactosidase that turns the color into blue once bound to its substrate (X-gal) as mentioned in figure (4) and graph (1).
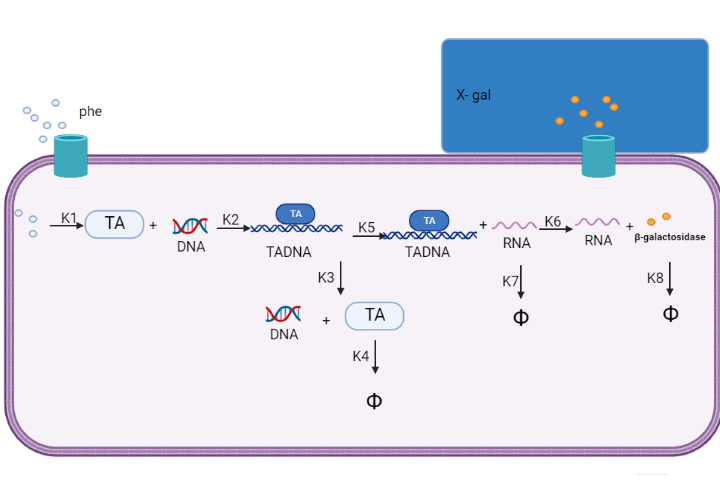
Figure (4) represents the cascade of reactions in whole cell-based biosensor model.
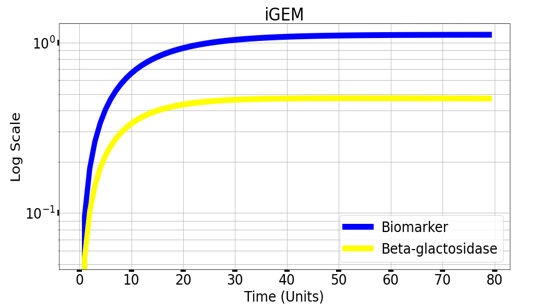
Graph(1) illustrates a direct relation between biomarker and beta-galactosidase ,so as the biomarker increases, the released amount of beta-galactosidase increases till it reaches constant value after about 30 time units. Therefore, the maximum amount of the biomarker releases the maximum amount of beta-galactosidase.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
[1] Yanbing Cui, Yiwei Meng, Juan Zhang,etc.Efficient secretory expression of recombinant proteins in Escherichia coli with a novel actinomycete signal peptide[J].Protein Expression and Purification,2017,129(1):69-74.
