Difference between revisions of "Part:BBa K274200"
| Line 20: | Line 20: | ||
For PDF version of this Datasheet: [[Image:Carotene.pdf]] | For PDF version of this Datasheet: [[Image:Carotene.pdf]] | ||
| + | |||
| + | |||
| + | '''Reference''' | ||
| + | |||
| + | Hal Alper, et al. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nature Biotechnology 23 (2005). | ||
| + | |||
| + | Nishizaki T, et al. Metabolic engineering of carotenoid biosynthesis in Escherichia coli by ordered gene assembly in Bacillus subtilis. Appl Environ Microbiol. 2007 Feb | ||
| + | |||
| + | Luke Z. Yuan, et al. Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metabolic Engineering 8 (2006). | ||
| + | |||
| + | Luan Tao, et al. Isolation of chromosomal mutations that affect carotenoid production in Escherichia coli: mutations alter copy number of ColE1-type plasmids. FEMS Microbiology Letters 243 (2005) | ||
| + | |||
| + | von Lintig J, et al. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving beta-carotene to retinal. J Biol Chem. 2000 Apr 21;275(16). | ||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 13:13, 21 October 2009
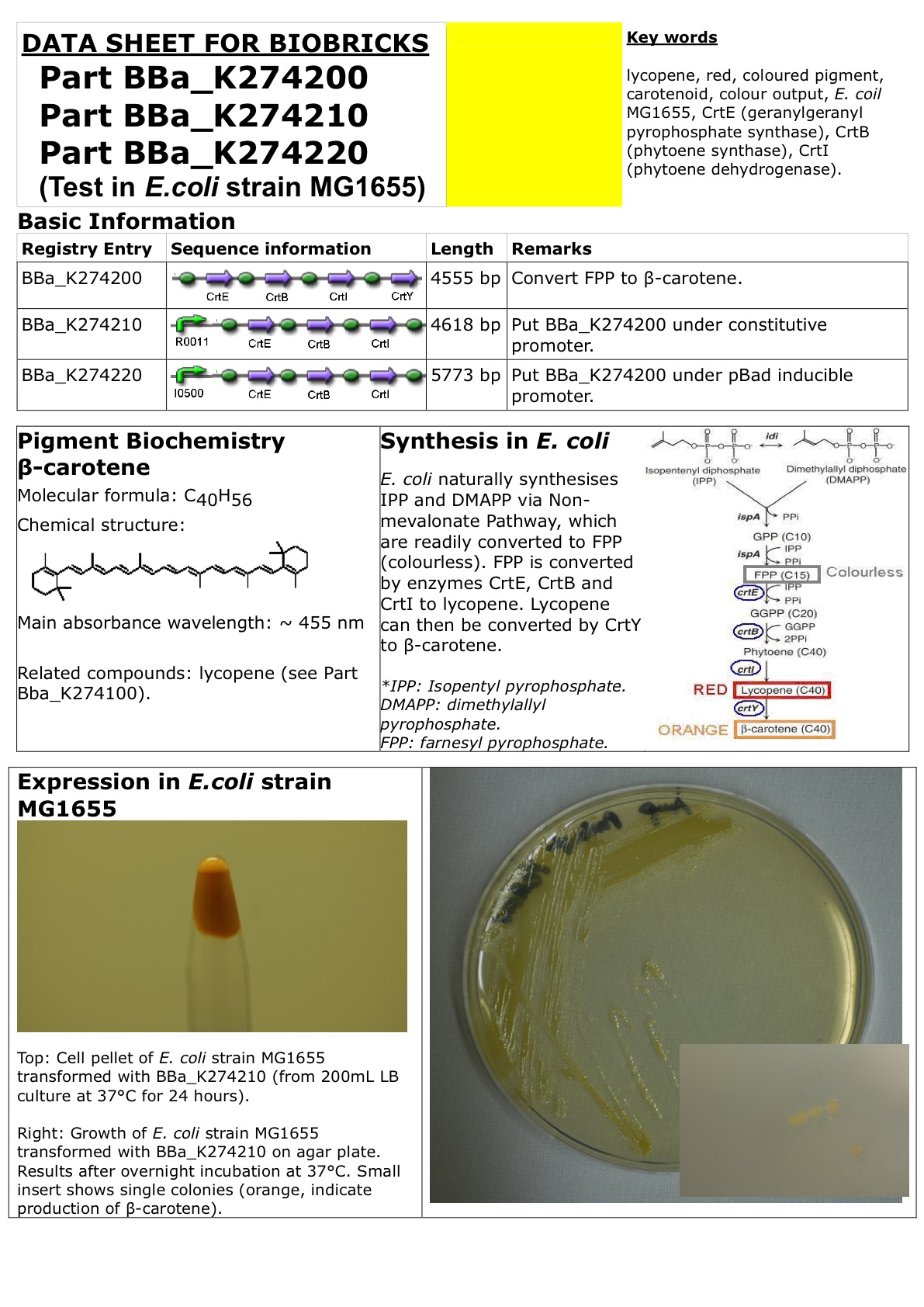
CrtEBIY with rbs
This Composite Biobrick is created by standard assembly of Part BBa_K118014 (CrtE with rbs), Part BBa_K118006 (CrtB with rbs), Part BBa_K118005 (CrtI with rbs) and Part BBa_K118013 (CrtY with rbs), which are submitted by previous iGEM teams. The whole cassette is on plasmid pSB1A2 (high copy, Ampicillin resistance).
Together, enzymes CrtE, CrtB, CrtI and CrtY convert colourless farnesyl pyrophosphate to orange beta-carotene (via intermediates geranylgeranyl pyroiphosphate, phytoene and lycopene). The orange beta-carotene pigment can then be used as a coloured signal output, e.g. for biosensors.
There is already individual ribosome binding site before each enzyme gene sequence. Internal restriction sites have been removed by previous iGEM teams. Please refer to Parts BBa_K118014, BBa_K118006, BBa_K118005 and BBa_K118013 for more information on individual Biobricks components.
This Composite Biobrick has been put under constitutive promoter R0011 (see Part BBa_K274210) and arabinose-inducible promoter I0500 (see Part BBa_274220), transformed and tested in E.coli strain MG1655.
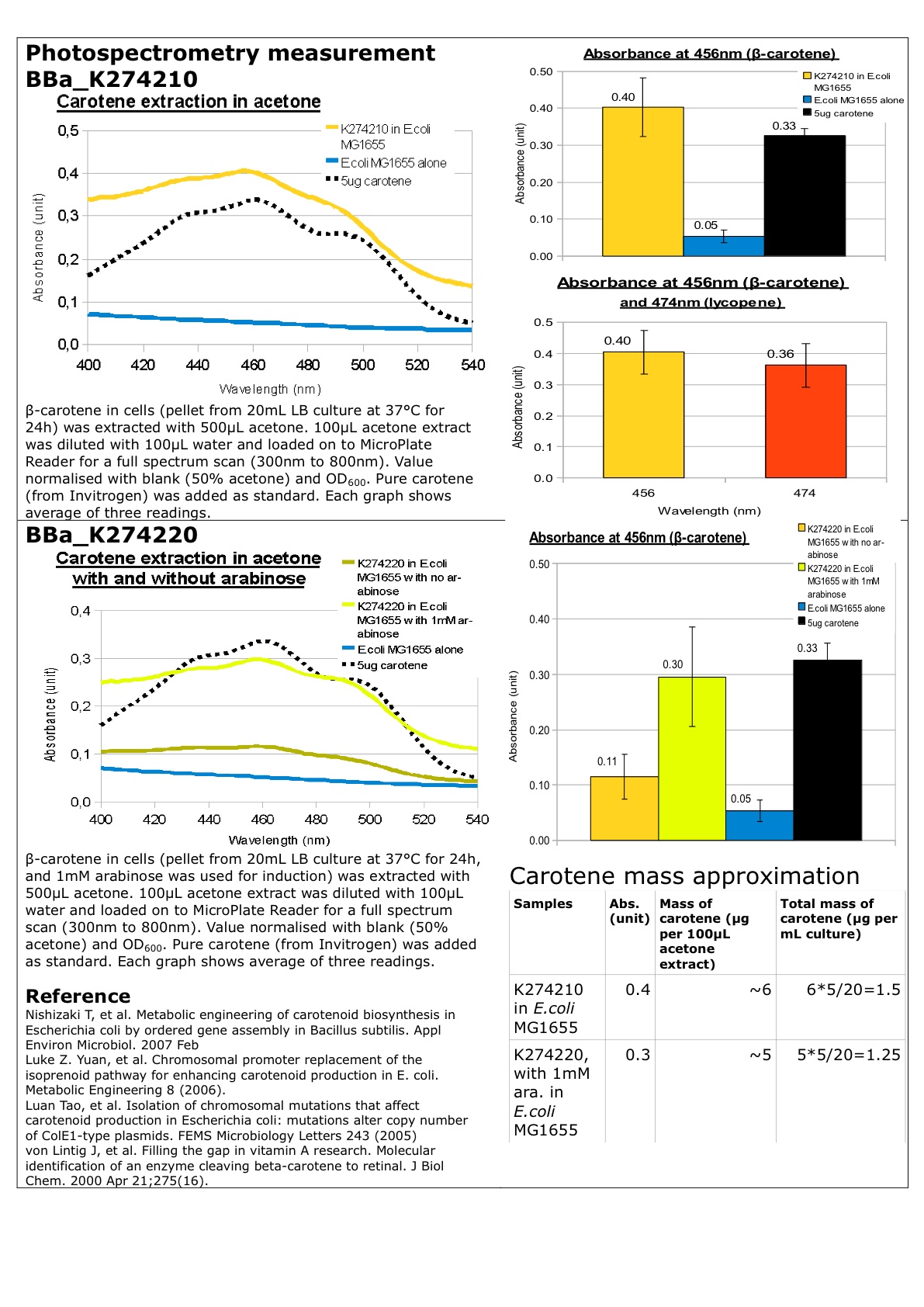
Amount of beta-carotene produced can be measured by photospectrometer with absorbance at 455nm (beta-carotene extraction using acetone).
A related Composite Biobrick is Part BBa_K274100 (CrtEBI with rbs), which "stops" at one step before beta-carotene, producing (and retaining) red lycopene pigment.
Datasheet for Part BBa_K274200, Part BBa_274210 and Part BBa_274220 in E. coli strain MG1655. You may also wish to refer to the "Experience" page.
For PDF version of this Datasheet: File:Carotene.pdf
Reference
Hal Alper, et al. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nature Biotechnology 23 (2005).
Nishizaki T, et al. Metabolic engineering of carotenoid biosynthesis in Escherichia coli by ordered gene assembly in Bacillus subtilis. Appl Environ Microbiol. 2007 Feb
Luke Z. Yuan, et al. Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metabolic Engineering 8 (2006).
Luan Tao, et al. Isolation of chromosomal mutations that affect carotenoid production in Escherichia coli: mutations alter copy number of ColE1-type plasmids. FEMS Microbiology Letters 243 (2005)
von Lintig J, et al. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving beta-carotene to retinal. J Biol Chem. 2000 Apr 21;275(16).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1974
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1510
Illegal NgoMIV site found at 1640
Illegal AgeI site found at 725 - 1000COMPATIBLE WITH RFC[1000]