|
|
| Line 25: |
Line 25: |
| | <!-- --> | | <!-- --> |
| | | | |
| − | ==iGEM 2020 QHFZ-China, new documentation (For Bronze)== | + | ==iGEM 2022 iBowu-China, new documentation (For Bronze)== |
| − | <h3><b>Group: QHFZ-China iGEM 2020</b></h3> | + | <h3><b>Group: iBowu-China iGEM 2022</b></h3> |
| − | <h3><b>Author: Yixian Yang</b></h3> | + | <h3><b>Author: Enshi Xv</b></h3> |
| − | <p> We measured [https://parts.igem.org/Part:BBa_J23100 BBa_J23100], [https://parts.igem.org/Part:BBa_J23107 BBa_J23107] and [https://parts.igem.org/Part:BBa_J23109 BBa_J23109] as a strong, moderate and weak promoter respectively in 2020. For all the experiments below, we use <i>E. coli</i> BL21(DE3) strain.</p> | + | <p> In this year’s iGEM competition, SHSBNU_China team focused on anthocyanins production, especially delphinidin. 4CL gene is one of the key gene in the synthetic pathway of delphinidin, which catalyzed phenylalanine into 4-coumaryl coenzyme A. </p> |
| − | <h3>Part 1: Measurement with a reprter, sfGFP</h3>
| + | We synthesized the sequence and constructed into plasmid pETDuet. </p> |
| − | <h4>Description</h4>
| + | After transforming to E.coli BL21 and induced by IPTG overnight at 16 degree Celsius at various concentrations (0.25 to 2 uM), we lysed the bacteria and performed SDS-PAGE and we confirmed 4CL enzyme to be expressed successfully. Judged by the thickness of the SDS-PAGE band width and darkness, concentrations of iPTG doesn’t play a critical role in the expression of 4CL enzyme. |
| − | <p> First, we measured the strength of the promoter by sfGFP [https://parts.igem.org/Part:BBa_K3457015 BBa_K3457015].</p>
| + | [[File:File:Bronze-1.jpg|600px|thumb|left|Figure 1. The expression test for BBa_K1033001]] |
| − | <h4>Protocol</h4>
| + | |
| − | <p> The gene circuit we used is as below:</p>
| + | |
| − | [[File:T--QHFZ-China--J2310-1.png|600px|thumb|left|Figure 1. The Schematic cartoon of the DNA construct to test J23100 /
| + | |
| − | J23107 / J23109 with sfGFP.]]
| + | |
| − | <p style="clear:left;"> The protocol is as below: <br>
| + | |
| − | (1) Pick clones which are in good condition and put them into 500 μL LB medium containing antibiotics. Shake them to
| + | |
| − | grow at 37℃ for 5~7 hours until the bacteria solution becomes turbid. <br>
| + | |
| − | (2) Add 2mM iPTG into 3 mL LB medium containing antibiotics. Add 3 μL of the bacteria solution mentioned in step 1
| + | |
| − | to dilute the bacteria by the ratio of 1:1000. Shake the solution to grow the bacteria at 37℃ overnight.<br>
| + | |
| − | (3) The bacteria solution was centrifuged and the LB medium was removed. Then the bacteria was resuspended by PBS.
| + | |
| − | 100 μL such solution was put into a well of a 96-well palte. The GFP fluorescence and OD<sub>600</sub> were detected
| + | |
| − | by a microplate readers (Bio-Teck). The parameters are: exciting light: 488 nm, light reception: 520 nm, gain: 50.
| + | |
| − | <br>
| + | |
| − | (4) The value of PBS was deducted from the result above. GFP / OD<sub>600</sub> was calculated.<br>
| + | |
| − | </p>
| + | |
| − | <h4>Result</h4>
| + | |
| − | [[File:T--QHFZ-China--sfGFP.jpg|600px|thumb|left|Figure 2. sfGFP was expressed with J23100 / J23107 / J23109.]]
| + | |
| − | <p style="clear:left;"> We set the strehgth of J23109 as 1. The relative strengths of J23107 and J23109 were 4.4 and
| + | |
| − | 12.0. Though they are not the same as the data at the top of this page, they worked well anb the strength order of
| + | |
| − | the three promoters was accordance was consistent with other people's data. The difference may owe to the certain
| + | |
| − | gene circuit and protocol. </p>
| + | |
| − | <h3>Part 2: Measurement with CHAS 106094</h3>
| + | |
| − | <h4>Description</h4>
| + | |
| − | <p> Second, we measured the strength of the promoter by CAHS 106094
| + | |
| − | [https://parts.igem.org/Part:BBa_K3457012 BBa_K3457012]. This year, we used CAHS 106094 to protect bacteria from
| + | |
| − | freeze-drying and dry storage. We used different promoters to adjust the expression level of CAHS 106094, to study
| + | |
| − | the relationship between the survival rate and CAHS 106094 expression level.</p>
| + | |
| − | <h4>Protocol</h4>
| + | |
| − | <p> The gene circuit we used is as below:</p>
| + | |
| − | [[File:T--QHFZ-China--J2310-2.png|600px|thumb|left|Figure 3. The Schematic cartoon of the DNA construct to test J23100 /
| + | |
| − | J23107 / J23109 with CAHS 106094.]]
| + | |
| − | <p style="clear:left;"> The protocol is as below: </p>
| + | |
| − | [[File:T--QHFZ-China--freeze-dry protocol.jpg|600px|thumb|left|Figure 4. Experiment protocol.]]
| + | |
| − | <p style="clear:left;">
| + | |
| − | 【Day 1】Induction culture<br>
| + | |
| − | (1) Pick clones which are in good condition and put them into 500 μL LB medium containing antibiotics. Shake them to
| + | |
| − | grow at 37℃ for 5~7 hours until the bacteria solution becomes turbid. <br>
| + | |
| − | (2) Add 2mM iPTG into 3 mL LB medium containing antibiotics. Add 3 μL of the bacteria solution mentioned in step 1
| + | |
| − | to dilute the bacteria by the ratio of 1:1000. Shake the solution to grow the bacteria at 37℃ overnight.<br>
| + | |
| − | 【Day 2】Freeze-dried<br>
| + | |
| − | (1) If fluorescence induced by the iPTG is detectable in the control group (GFP), continue conducting the
| + | |
| − | experiment.<br>
| + | |
| − | (2) Use spectrophotometer to measure the OD<sub>600</sub> of the bacteria solution, OD<sub>600</sub> = 1 equals to
| + | |
| − | 10<sup>9</sup> cells. If the OD<sub>600</sub> value is between 0.1 and 1, There is a linear relationship between
| + | |
| − | OD<sub>600</sub> and bacterial density. Calculate the volume of bacterial solution for 10<sup>9</sup> cells by using
| + | |
| − | the formula V = 100 / (OD<sub>600</sub> × Dilution ratio).<br>
| + | |
| − | (3) Take out a measured amount of 10<sup>9</sup> cells and centrifuge it at 8000 rpm for 3 min. Then pour out the
| + | |
| − | supernatant.<br>
| + | |
| − | (4) Resuspend the bacteria in a 15 mL tube with pre-refrigerated 100 μL 3% glucose solution.<br>
| + | |
| − | (5) Take off the cover of the tube and put the bacteria into the cold trap. Open the compressor of the
| + | |
| − | lyophilization machine and freeze the shake tube for 2 h at -70℃.<br>
| + | |
| − | (6) Put the caky bacteria solution into the drying chamber of the lyophilization machine. Open the vacuum pump to
| + | |
| − | dry it in vacuum for 6h at 1 Pa vacuum degree.<br>
| + | |
| − | (7) Turn off the vacuum pump, place it at seal box filled with silica-gel desiccant a for 2 days at room
| + | |
| − | temperature.<br>
| + | |
| − | 【Day 3】Room temperature storage<br>
| + | |
| − | 【Day 4】Detect the survival rate<br>
| + | |
| − | (1) Add 1 mL of sterile water to the tube, vortex for 15 s, placed it at room temperature for 10 min.<br>
| + | |
| − | (2) Adjust the density of the bacteria solution by gradient dilution, then spread 100 μL of the bacteria solution on
| + | |
| − | the LB plate.<br>
| + | |
| − | (3) If the density above is not suitable, take 100μL of the solution and spread it on the LB plate after several
| + | |
| − | gradient dilutions.<br>
| + | |
| − | (4) Culture the bacteria overnight at 37℃.<br>
| + | |
| − | 【Day 5】Cell Count<br>
| + | |
| − | (1) Take out the LB plate and take photos to record experimental results.<br>
| + | |
| − | (2) Use the automatic cell counting function of Image J to count the colone number on the LB plate, then compare the
| + | |
| − | results between each group.<br>
| + | |
| − | </p>
| + | |
| − | <h4>Result</h4>
| + | |
| − | [[File:T--QHFZ-China--J2310-3.png|600px|thumb|left|Figure 5. The Cfu of bacteria expressing CAHS 106094 after | + | |
| − | freeze-drying with J23100 / J23107 / J23109.]]
| + | |
| − | <p style="clear:left;"> As expected, J23100 is the strongest promoter and it gave the best survival rate. J23107 is
| + | |
| − | the second and J23109 seemed too weak to express enough CAHS 106094. In conclusion, J23100 and J23107 is effective
| + | |
| − | in this situation, but J23109 is not.</p>
| + | |
| | <!-- The end of QHFZ-China 2020--> | | <!-- The end of QHFZ-China 2020--> |
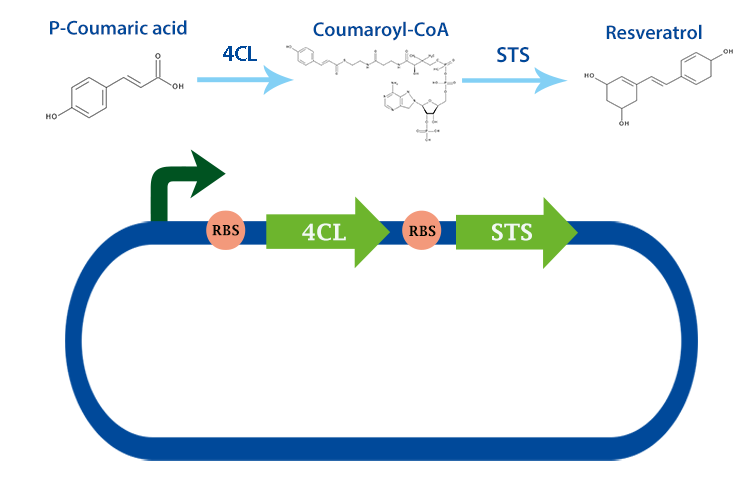
4-coumarate ligase (4CL) is an enzyme that catalyses the reaction from p-coumaric acid to 4-coumaroyl-coenzyme A (4-coumaryl-CoA). This enzyme is derived from the plant arabidhopsis thaliana, but exists in many other plants. [1]
In our project, we have been using it together with stilbene synthase, that produces resveratrol with the help of this enzyme.
In this year’s iGEM competition, SHSBNU_China team focused on anthocyanins production, especially delphinidin. 4CL gene is one of the key gene in the synthetic pathway of delphinidin, which catalyzed phenylalanine into 4-coumaryl coenzyme A.
We synthesized the sequence and constructed into plasmid pETDuet. </p>
After transforming to E.coli BL21 and induced by IPTG overnight at 16 degree Celsius at various concentrations (0.25 to 2 uM), we lysed the bacteria and performed SDS-PAGE and we confirmed 4CL enzyme to be expressed successfully. Judged by the thickness of the SDS-PAGE band width and darkness, concentrations of iPTG doesn’t play a critical role in the expression of 4CL enzyme.