Difference between revisions of "Part:BBa K4162005"
| Line 8: | Line 8: | ||
Hammerhead ribozyme was first found in the genome of viruses and viroids. It involved in the processing of RNA transcripts based on rolling-circle replication. The tandem copy of RNA sequence will be generated in the roll ring replication, and the self-cleaving activity of ribozyme can ensure the generation of RNA copy of unit length.<ref>Ferré-D'Amaré, Adrian R, and William G Scott. “Small self-cleaving ribozymes.” Cold Spring Harbor perspectives in biology vol. 2,10 (2010): a003574. doi:10.1101/cshperspect.a003574</ref> | Hammerhead ribozyme was first found in the genome of viruses and viroids. It involved in the processing of RNA transcripts based on rolling-circle replication. The tandem copy of RNA sequence will be generated in the roll ring replication, and the self-cleaving activity of ribozyme can ensure the generation of RNA copy of unit length.<ref>Ferré-D'Amaré, Adrian R, and William G Scott. “Small self-cleaving ribozymes.” Cold Spring Harbor perspectives in biology vol. 2,10 (2010): a003574. doi:10.1101/cshperspect.a003574</ref> | ||
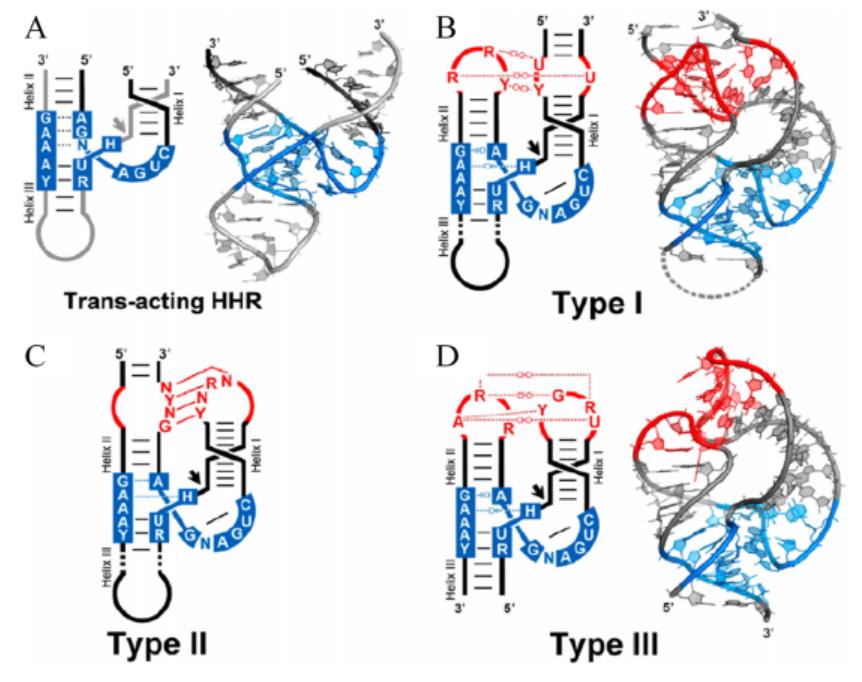
| − | The secondary structure of hammerhead ribozyme resembles a hammer. According to different open helix tips, hammerhead ribozymes can be divided into three types: TypeⅠ, Type II and Type III. The catalytic center of ribozyme consists of 15 highly conserved bases surrounded by three helixs(HelixⅠ, Helix II and Helix III). The long-range interaction between HelixⅠ and Helix II can help stabilize the conformation of the catalytic center of the enzyme and improve the catalytic efficiency. | + | The secondary structure of hammerhead ribozyme resembles a hammer. According to different open helix tips, hammerhead ribozymes can be divided into three types: TypeⅠ, Type II and Type III. The catalytic center of ribozyme consists of 15 highly conserved bases surrounded by three helixs(HelixⅠ, Helix II and Helix III). The long-range interaction between HelixⅠ and Helix II can help stabilize the conformation of the catalytic center of the enzyme and improve the catalytic efficiency (Figure 1).<ref>Jimenez, Randi M et al. “Chemistry and Biology of Self-Cleaving Ribozymes.” Trends in biochemical sciences vol. 40,11 (2015): 648-661. doi:10.1016/j.tibs.2015.09.001</ref> |
[[File:T--Fudan--hammerhead ribozyme.png|400px|thumb|none|'''Figure 1. Overall tertiary structure of different types of hammerhead ribozymes.'''Blue marks highly conserved bases. The black arrow marks the digestion site. Red marks the long-range interaction between HelixⅠand Helix II.]] | [[File:T--Fudan--hammerhead ribozyme.png|400px|thumb|none|'''Figure 1. Overall tertiary structure of different types of hammerhead ribozymes.'''Blue marks highly conserved bases. The black arrow marks the digestion site. Red marks the long-range interaction between HelixⅠand Helix II.]] | ||
Revision as of 09:14, 6 October 2022
Hammerhead ribozyme
Introduction
Hammerhead ribozyme was first found in the genome of viruses and viroids. It involved in the processing of RNA transcripts based on rolling-circle replication. The tandem copy of RNA sequence will be generated in the roll ring replication, and the self-cleaving activity of ribozyme can ensure the generation of RNA copy of unit length.[1]
The secondary structure of hammerhead ribozyme resembles a hammer. According to different open helix tips, hammerhead ribozymes can be divided into three types: TypeⅠ, Type II and Type III. The catalytic center of ribozyme consists of 15 highly conserved bases surrounded by three helixs(HelixⅠ, Helix II and Helix III). The long-range interaction between HelixⅠ and Helix II can help stabilize the conformation of the catalytic center of the enzyme and improve the catalytic efficiency (Figure 1).[2]
Usage and Biology
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
- ↑ Ferré-D'Amaré, Adrian R, and William G Scott. “Small self-cleaving ribozymes.” Cold Spring Harbor perspectives in biology vol. 2,10 (2010): a003574. doi:10.1101/cshperspect.a003574
- ↑ Jimenez, Randi M et al. “Chemistry and Biology of Self-Cleaving Ribozymes.” Trends in biochemical sciences vol. 40,11 (2015): 648-661. doi:10.1016/j.tibs.2015.09.001